
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.

The goal of bolasso is to implement model-consistent Lasso estimation via the bootstrap [1].
You can install the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("dmolitor/bolasso")To illustrate the usage of bolasso, we’ll use the Pima Indians Diabetes dataset to determine which factors are important predictors of testing positive for diabetes. For a full description of the input variables, see the link above.
library(bolasso)
data(PimaIndiansDiabetes, package = "mlbench")
# Quick overview of the dataset
str(PimaIndiansDiabetes)
#> 'data.frame': 768 obs. of 9 variables:
#> $ pregnant: num 6 1 8 1 0 5 3 10 2 8 ...
#> $ glucose : num 148 85 183 89 137 116 78 115 197 125 ...
#> $ pressure: num 72 66 64 66 40 74 50 0 70 96 ...
#> $ triceps : num 35 29 0 23 35 0 32 0 45 0 ...
#> $ insulin : num 0 0 0 94 168 0 88 0 543 0 ...
#> $ mass : num 33.6 26.6 23.3 28.1 43.1 25.6 31 35.3 30.5 0 ...
#> $ pedigree: num 0.627 0.351 0.672 0.167 2.288 ...
#> $ age : num 50 31 32 21 33 30 26 29 53 54 ...
#> $ diabetes: Factor w/ 2 levels "neg","pos": 2 1 2 1 2 1 2 1 2 2 ...First, we run 100-fold bootstrapped Lasso with the
glmnet implementation. We can get a rough estimate of the
elapsed time using system.time().
system.time({
model <- bolasso(
diabetes ~ .,
data = PimaIndiansDiabetes,
n.boot = 100,
implement = "glmnet",
family = "binomial"
)
})
#> Loaded glmnet 4.1-3
#> user system elapsed
#> 19.32 0.14 19.58We can get a quick overview of the model by printing the
bolasso object.
model
#> ------------- 100-fold bootstrapped Lasso -------------
#>
#> Model matrix dimensions:
#> - 8 Predictors
#> - 768 Observations
#>
#> Selected variables:
#> - 6/8 predictors selected with 90% threshold
#> - 4/8 predictors selected with 100% thresholdNext, we can extract all variables that were selected in 90% and 100%
of the bootstrapped Lasso models. We can also pass any relevant
arguments to predict on the cv.glmnet or
cv.gamlr model objects. In this case we will use the lambda
value that minimizes OOS error.
selected_vars(model, threshold = 0.9, select = "lambda.min")
#> # A tibble: 7 x 2
#> variable mean_coef
#> <chr> <dbl>
#> 1 Intercept -8.15
#> 2 pregnant 0.119
#> 3 glucose 0.0348
#> 4 pressure -0.0113
#> 5 mass 0.0821
#> 6 pedigree 0.849
#> 7 age 0.0138
selected_vars(model, threshold = 1, select = "lambda.min")
#> # A tibble: 5 x 2
#> variable mean_coef
#> <chr> <dbl>
#> 1 Intercept -8.15
#> 2 pregnant 0.119
#> 3 glucose 0.0348
#> 4 mass 0.0821
#> 5 pedigree 0.849We can also quickly plot the selected variables at the 90% and 100% threshold values.
plot(model, threshold = 0.9)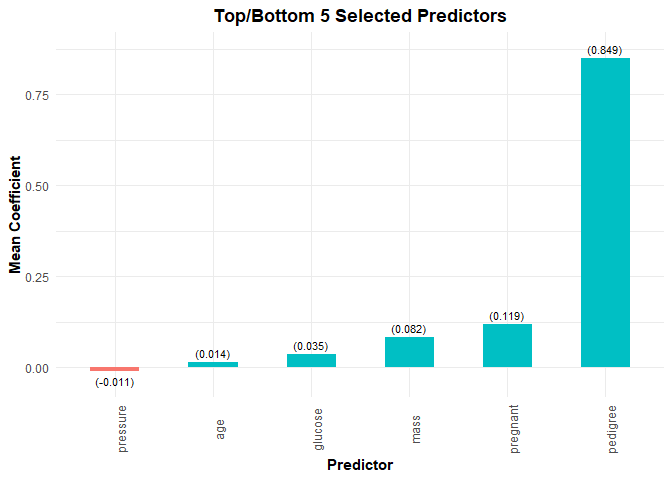
plot(model, threshold = 1)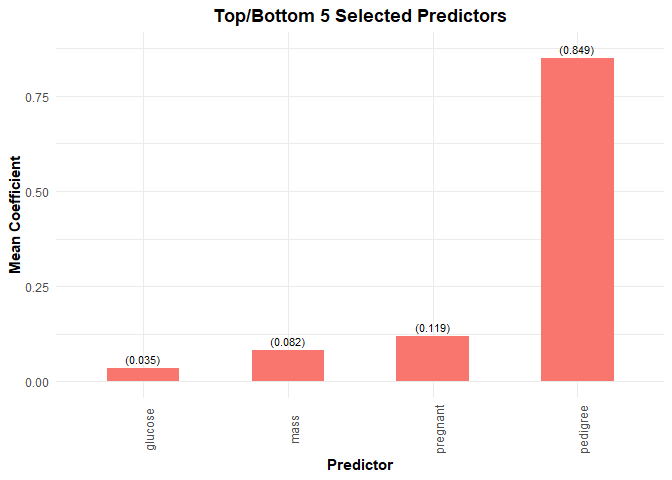
We can execute bolasso in parallel via the future package. To
do so we can copy the code from above with only one minor tweak shown
below.
future::plan("multisession")We can now run the code from above, unaltered, and it will execute in parallel.
system.time({
model <- bolasso(
diabetes ~ .,
data = PimaIndiansDiabetes,
n.boot = 100,
implement = "glmnet",
family = "binomial"
)
})
#> user system elapsed
#> 0.17 0.03 5.58[1] Bach, Francis. “Bolasso: Model Consistent Lasso Estimation through the Bootstrap.” ArXiv:0804.1302 [Cs, Math, Stat], April 8, 2008. https://arxiv.org/abs/0804.1302.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.