
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.

R package for Airborne LiDAR Data Manipulation and Visualization for Forestry Applications
The lidR package provides functions to read and write
.las and .laz files, plot point clouds,
compute metrics using an area-based approach, compute digital canopy
models, thin LiDAR data, manage a collection of LAS/LAZ files,
automatically extract ground inventories, process a collection of tiles
using multicore processing, segment individual trees, classify points
from geographic data, and provides other tools to manipulate LiDAR data
in a research and development context.
lidR from R with:
install.packages("lidR")lidR. It is free and open source, but requires time and
effort to develop and maintain.lidR has been cited by more than 1,000 scientific
papers. To cite the package use citation() from within
R:
citation("lidR")
#> Roussel, J.R., Auty, D., Coops, N. C., Tompalski, P., Goodbody, T. R. H., Sánchez Meador, A., Bourdon, J.F., De Boissieu, F., Achim, A. (2021). lidR : An R package for analysis of Airborne Laser Scanning (ALS) data. Remote Sensing of Environment, 251 (August), 112061. <doi:10.1016/j.rse.2020.112061>.
#> Jean-Romain Roussel and David Auty (2023). Airborne LiDAR Data Manipulation and Visualization for Forestry Applications. R package version 3.1.0. https://cran.r-project.org/package=lidRYou may also be interested by our new lasR
package.
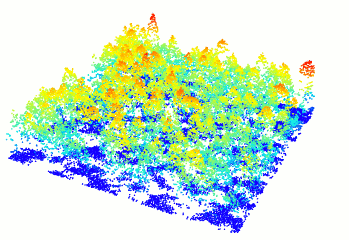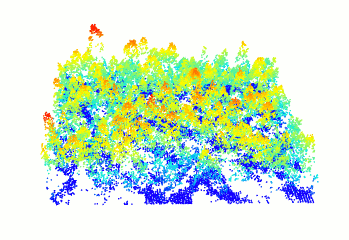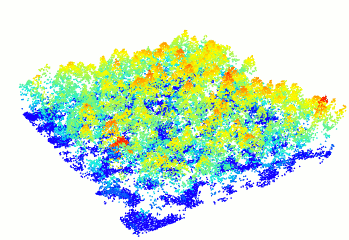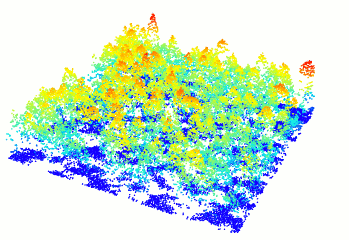
In R-fashion style the function plot, based on
rgl, enables the user to display, rotate and zoom a point
cloud.
las <- readLAS("<file.las>")
plot(las)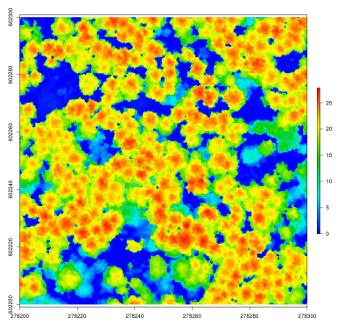
lidR has several algorithms from the literature to
compute canopy height models either point-to-raster based or
triangulation based. This allows testing and comparison of some methods
that rely on a CHM, such as individual tree segmentation or the
computation of a canopy roughness index.
las <- readLAS("<file.las>")
# Khosravipour et al. pitfree algorithm
thr <- c(0,2,5,10,15)
edg <- c(0, 1.5)
chm <- rasterize_canopy(las, 1, pitfree(thr, edg))
plot(chm)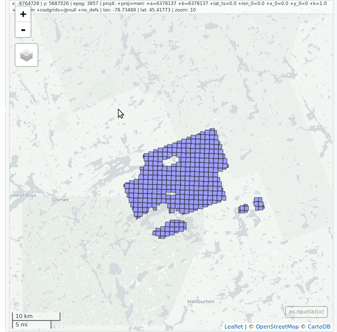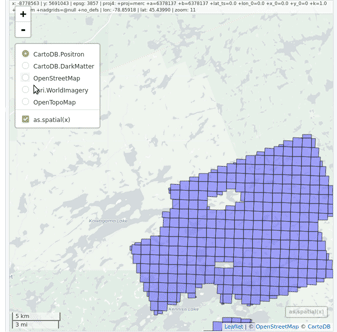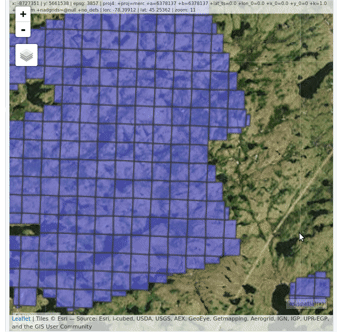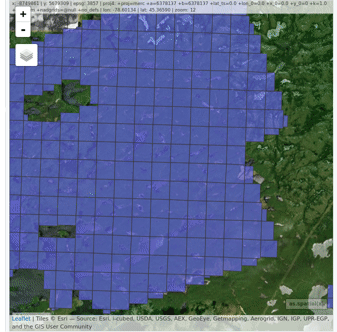
lidR enables the user to manage, use and process a
collection of las files. The function
readLAScatalog builds a LAScatalog object from
a folder. The function plot displays this collection on an
interactive map using the mapview package (if
installed).
ctg <- readLAScatalog("<folder/>")
plot(ctg, map = TRUE)From a LAScatalog object the user can (for example)
extract some regions of interest (ROI) with clip_roi().
Using a catalog for the extraction of the ROI guarantees fast and
memory-efficient clipping. LAScatalog objects allow many
other manipulations that can be done with multicore processing.
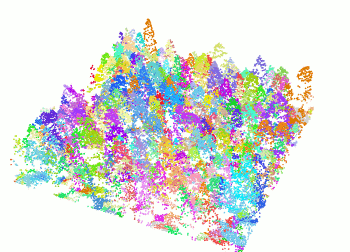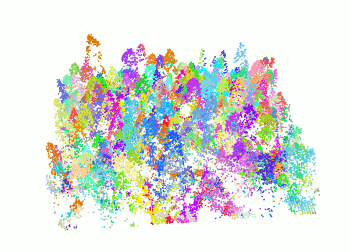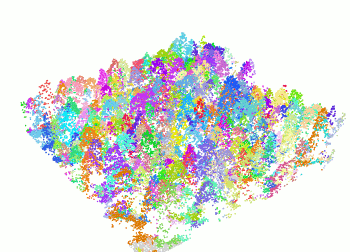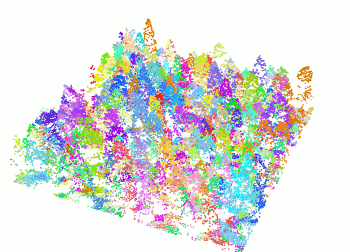
The segment_trees() function has several algorithms from
the literature for individual tree segmentation, based either on the
digital canopy model or on the point-cloud. Each algorithm has been
coded from the source article to be as close as possible to what was
written in the peer-reviewed papers. Our goal is to make published
algorithms usable, testable and comparable.
las <- readLAS("<file.las>")
las <- segment_trees(las, li2012())
col <- random.colors(200)
plot(las, color = "treeID", colorPalette = col)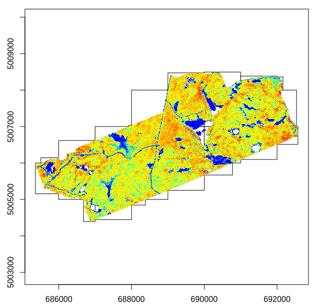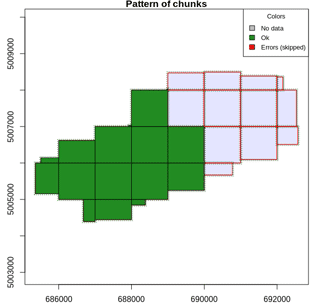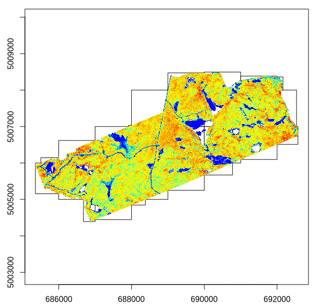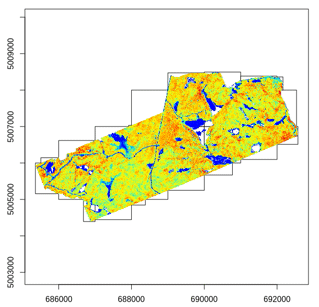
Most of the lidR functions can seamlessly process a set of tiles and
return a continuous output. Users can create their own methods using the
LAScatalog processing engine via the
catalog_apply() function. Among other features the engine
takes advantage of point indexation with lax files, takes care of
processing tiles with a buffer and allows for processing big files that
do not fit in memory.
# Load a LAScatalog instead of a LAS file
ctg <- readLAScatalog("<path/to/folder/>")
# Process it like a LAS file
chm <- rasterize_canopy(ctg, 2, p2r())
col <- random.colors(50)
plot(chm, col = col)
lidR can read full waveform data from LAS files and provides interpreter functions to convert the raw data into something easier to manage and display in R. The support of FWF is still in the early stages of development.
fwf <- readLAS("<fullwaveform.las>")
# Interpret the waveform into something easier to manage
las <- interpret_waveform(fwf)
# Display discrete points and waveforms
x <- plot(fwf, colorPalette = "red", bg = "white")
plot(las, color = "Amplitude", add = x)lidR is developed openly by r-lidar.
The development of lidR was made possible through the
financial support of Laval
University, the AWARE project
and Ministry
of Natural Ressources and Forests of Québec. To continue the
development of this free software, we now offer consulting, programming,
and training services. For more information, please visit our website.
# Ubuntu
sudo add-apt-repository ppa:ubuntugis/ubuntugis-unstable
sudo apt-get update
sudo apt-get install libgdal-dev libgeos++-dev libudunits2-dev libproj-dev libx11-dev libgl1-mesa-dev libglu1-mesa-dev libfreetype6-dev libxt-dev libfftw3-dev
# Fedora
sudo dnf install gdal-devel geos-devel udunits2-devel proj-devel mesa-libGL-devel mesa-libGLU-devel freetype-devel libjpeg-turbo-develThese binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.