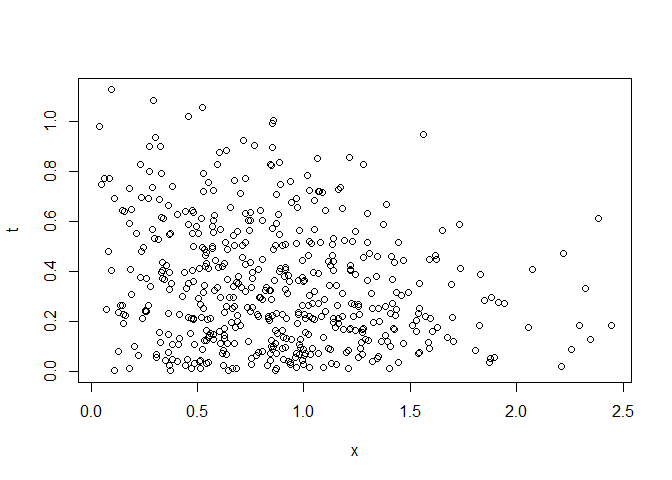
Fig.1. Random data of biomarker and time-to-event
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
The goal of parTimeROC is to store methods and procedures needed to run the time-dependent ROC analysis parametrically. This package adopts two different theoretical framework to produce the ROC curve which are from the proportional hazard model and copula function. Currently, this package only able to run analysis for single covariate/biomarker with survival time. The future direction for this work is to be able to include analysis for multiple biomarkers with longitudinal measurements.
You can install the development version of parTimeROC from GitHub with:
# install.packages("devtools")
devtools::install_github("FaizAzhar/parTimeROC")Since this package also include the bayesian estimation procedure (rstan), please ensure to follow the correct installation setup such as demonstrated in this article.
A receiver operating characteristics (ROC) curve is a curve that measures a model’s accuracy to correctly classify a population into a binary status (eg: dead/alive). The curve acts as a tool for analysts to compare which model is suitable to be used as a classifiers. However, in survival analysis, it is noted that the status of population fluctuate across time. Thus, a standard ROC analysis might underestimates the true accuracy measurement that the classification model have. In a situation where the population might enter or exit any of the two status over time, including the time component into the ROC analysis is shown to be superior and can help analysts to assess the performance of the model’s accuracy over time. In addition, a time-dependent ROC can also show at which specific time point a model will have a similar performance measurement with other model.
For the time being, two methods are frequently used when producing the time-dependent ROC curve. The first method employs the Cox proportional hazard model (PH) to estimate the joint distribution of the covariates and time-to-event. The second method employs a copula function which link the marginal distributions of covariates and time-to-event to estimate its joint distribution. After obtaining estimates for the joint distribution, two metrics can be computed which is the time-dependent sensitivity and specificity. Plotting these two informations will generate the desired time-dependent ROC curve.
Explanations below are showing the functions that can be found within
parTimeROC package and its implementation.
timeroc_objFollowing an OOP approaches, a TimeROC object can be
initialized by using the parTimeROC::timeroc_obj()
method.
test <- parTimeROC::timeroc_obj("normal-gompertz-PH")
print(test)
#> Model Assumptions: Proportional Hazard (PH)
#> X : Gaussian
#> Time-to-Event : Gompertz
test <- parTimeROC::timeroc_obj("normal-gompertz-copula", copula = "gumbel90")
print(test)
#> Model Assumptions: 90 Degrees Rotated Gumbel Copula
#> X : Gaussian
#> Time-to-Event : GompertzNotice that we included the print method to generate the summary for
the test object which has a TimeROC class.
A list of distributions and copula have been stored within this
package. It is accessible via the get.distributions or
get.copula script.
names(parTimeROC::get.distributions)
#> [1] "exponential" "weibull" "gaussian" "normal" "lognormal"
#> [6] "gompertz" "skewnormal"
names(parTimeROC::get.copula)
#> [1] "gaussian" "clayton90" "gumbel90" "gumbel" "joe90"rtimerocCommon tasks in mathematical modelling are prepared. For simulation
purposes, procedure to generate random data from PH or copula function
is created. The random data can be obtained using the
parTimeROC::rtimeroc(). The
parTimeROC::rtimeroc() returns a dataframe of 3 columns (t,
x, status).
library(parTimeROC)
## PH model
test <- timeroc_obj(dist = 'weibull-gompertz-PH')
set.seed(23456)
rr <- rtimeroc(obj = test, censor.rate = 0.5, n=500,
params.t = c(shape=2, rate=1),
params.x = c(shape=2, scale=1),
params.ph=0.5)
plot(t~x, rr)
Fig.1. Random data of biomarker and time-to-event
timeroc_fitWe can also fit datasets that have time-to-event, covariates and
status columns with the PH or copula model using the
parTimeROC::timeroc_fit().
For PH model, two fitting processes are done. One is to fit the biomarker distribution alone. Another is to fit the time-to-event that is assumed to follow a proportional hazard model.
Meanwhile, for copula method, the IFM technique is used due to its light computational requirement. Three fitting processes are conducted. One is to fit the marginal distribution for biomarker, another is to fit the marginal time-to-event. And lastly is to fit the copula function.
User can choose to conduct the model fitting procedure based on the
frequentist or bayesian approach by specifying the
method = 'mle' or method = 'bayes' within the
parTimeROC::timeroc_fit() function.
By default, the frequentist approach is used to estimate the model’s parameters.
library(parTimeROC)
## fitting copula model
test <- timeroc_obj(dist = 'gompertz-gompertz-copula', copula = "gumbel90")
set.seed(23456)
rr <- rtimeroc(obj = test, censor.rate = 0, n=500,
params.t = c(shape=3,rate=1),
params.x = c(shape=1,rate=2),
params.copula=-5) # name of parameter must follow standard
cc <- timeroc_fit(rr$x, rr$t, rr$event, obj = test)
print(cc)
#> Model: gompertz-gompertz-copula
#> ------
#> X (95% CI) :
#> AIC = -65.51402
#> est low upper se
#> shape 0.9343 0.6216 1.2469 0.1595
#> rate 2.0931 1.7975 2.3887 0.1508
#> ------
#> Time-to-Event (95% CI) :
#> AIC = -141.7148
#> est low upper se
#> shape 3.0894 2.7066 3.4722 0.1953
#> rate 0.9160 0.7547 1.0774 0.0823
#> ------
#> Copula (95% CI) :
#> AIC = -1432.074
#> est low upper se
#> theta -5.1126 -5.4868 -4.7384 0.1909Notice that the print method also can be used to print the results obtained from the fitting process.
timeroc_gofAfter fitting the model with either PH or copula model, its
goodness-of-fit can be examined through the function
parTimeROC::timeroc_gof(). This will return a list of test
statistic and p-values denoting misspecification of model or not.
Kolmogorov-Smirnov testing is performed for model checking. If
p-value < 0.05, we reject the null hypothesis that the
data (biomarker or time-to-event) are following the assumed
distribution.
For copula model, additional testing is needed to check whether the
copula used is able to model the data or not. After using the Rosenblatt
transformation, we conduct an independent testing to check whether the
empirical conditional and cumulative distribution are independent. If
the p-value < 0.05, we reject the null hypothesis which
stated that the conditional and cumulative are independent. Thus, for
p-value < 0.05, the copula failed to provide a good
estimation for the joint distribution.
library(parTimeROC)
# Copula model
rt <- timeroc_obj("normal-weibull-copula",copula="clayton90")
set.seed(1)
rr <- rtimeroc(rt, n=300, censor.rate = 0,
params.x = c(mean=5, sd=1),
params.t = c(shape=1, scale=5),
params.copula = -2.5)
test <- timeroc_obj("normal-weibull-copula",copula="gumbel90")
jj <- timeroc_fit(test, rr$x, rr$t, rr$event)
timeroc_gof(jj)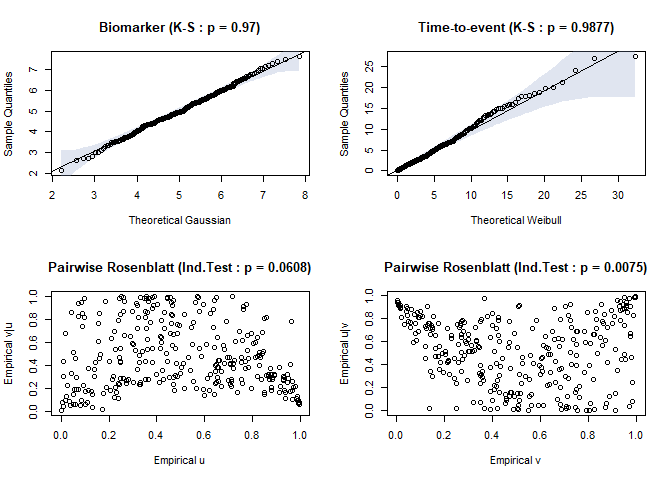
Fig.2. Residual plots for biomarker and time-to-event distribution when misspecified
#> $ks_x
#>
#> Asymptotic two-sample Kolmogorov-Smirnov test
#>
#> data: df$x and theo.q
#> D = 0.04, p-value = 0.97
#> alternative hypothesis: two-sided
#>
#>
#> $ks_t
#>
#> Asymptotic two-sample Kolmogorov-Smirnov test
#>
#> data: df$t and theo.q
#> D = 0.036667, p-value = 0.9877
#> alternative hypothesis: two-sided
#>
#>
#> $ind_u
#> $ind_u$statistic
#> [1] 1.875196
#>
#> $ind_u$p.value
#> [1] 0.06076572
#>
#>
#> $ind_v
#> $ind_v$statistic
#> [1] 2.674574
#>
#> $ind_v$p.value
#> [1] 0.007482435test <- timeroc_obj("normal-weibull-copula",copula="clayton90")
jj <- timeroc_fit(test, rr$x, rr$t, rr$event)
timeroc_gof(jj)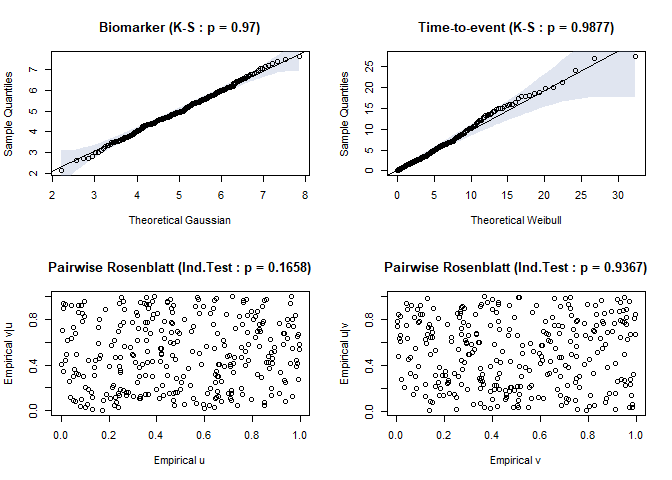
Fig.3. Residual plots for biomarker and time-to-event distribution when correct specification
#> $ks_x
#>
#> Asymptotic two-sample Kolmogorov-Smirnov test
#>
#> data: df$x and theo.q
#> D = 0.04, p-value = 0.97
#> alternative hypothesis: two-sided
#>
#>
#> $ks_t
#>
#> Asymptotic two-sample Kolmogorov-Smirnov test
#>
#> data: df$t and theo.q
#> D = 0.036667, p-value = 0.9877
#> alternative hypothesis: two-sided
#>
#>
#> $ind_u
#> $ind_u$statistic
#> [1] 1.385664
#>
#> $ind_u$p.value
#> [1] 0.1658495
#>
#>
#> $ind_v
#> $ind_v$statistic
#> [1] 0.07947699
#>
#> $ind_v$p.value
#> [1] 0.9366532library(parTimeROC)
# PH model
rt <- timeroc_obj("normal-weibull-PH")
set.seed(1)
rr <- rtimeroc(rt, n=300, censor.rate = 0,
params.x = c(mean=5, sd=1),
params.t = c(shape=1, scale=5),
params.ph = 1.2)
test <- timeroc_obj("lognormal-lognormal-PH")
jj <- timeroc_fit(test, rr$x, rr$t, rr$event)
timeroc_gof(jj)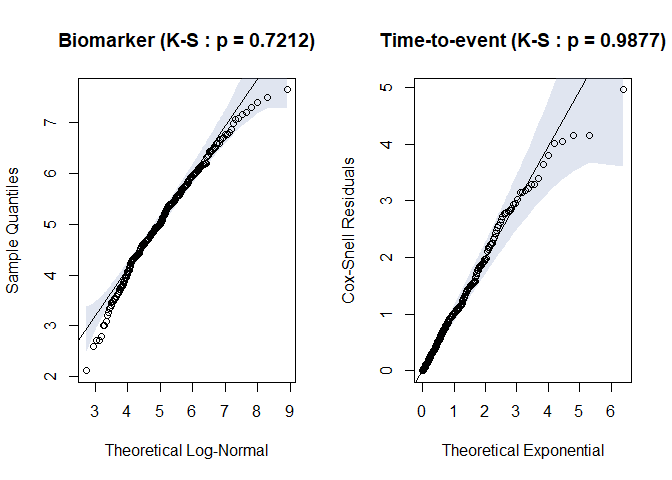
Fig.4. Residual plots for biomarker and time-to-event distribution when misspecified
#> $ks_x
#>
#> Asymptotic two-sample Kolmogorov-Smirnov test
#>
#> data: df$x and theo.q
#> D = 0.056667, p-value = 0.7212
#> alternative hypothesis: two-sided
#>
#>
#> $ks_t
#>
#> Asymptotic two-sample Kolmogorov-Smirnov test
#>
#> data: df$coxsnell and theo.q
#> D = 0.036667, p-value = 0.9877
#> alternative hypothesis: two-sidedtest <- timeroc_obj("normal-weibull-PH")
jj <- timeroc_fit(test, rr$x, rr$t, rr$event)
timeroc_gof(jj)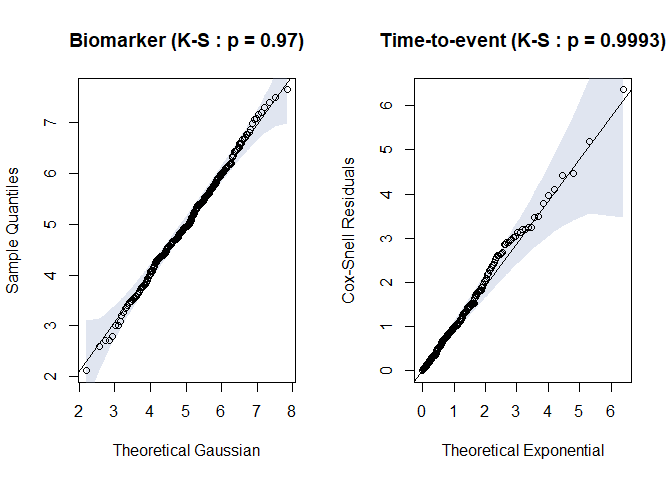
Fig.5. Residual plots for biomarker and time-to-event distribution when correct specification
#> $ks_x
#>
#> Asymptotic two-sample Kolmogorov-Smirnov test
#>
#> data: df$x and theo.q
#> D = 0.04, p-value = 0.97
#> alternative hypothesis: two-sided
#>
#>
#> $ks_t
#>
#> Asymptotic two-sample Kolmogorov-Smirnov test
#>
#> data: df$coxsnell and theo.q
#> D = 0.03, p-value = 0.9993
#> alternative hypothesis: two-sidedtimeroc_predictFinally, after fitting process, we can predict the value of
sensitivity and specificity of the covariates at specific time point
using the parTimeROC::timeroc_predict() function. This will
return a list of dataframe for each specified time.
To generate the ROC curve, user can choose to conduct the prediction
procedure using the type = 'standard' or
type = 'landmark' approach.
By default, the type = 'standard' analysis will be used
to produce the ROC curve at different time point. After model fitting
procedure, the estimated parameters will be extracted and used to
compute the ROC at the specified time of interest.
Meanwhile for the type = 'landmark' analysis, at each
time point of interest, the status of each observation will be updated
prior running the model fitting procedure. Hence, in landmark analysis,
the fitting procedure will be conducted multiple times. At each time of
interest, the updated estimators are then used to produce the ROC
curve.
library(parTimeROC)
# Copula model
test <- timeroc_obj(dist = 'gompertz-gompertz-copula', copula='clayton90',
params.t = c(shape=3,rate=1),
params.x = c(shape=1,rate=2),
params.copula=-5)
set.seed(23456)
rr <- rtimeroc(obj = test, censor.rate = 0.2, n=500)
cc <- timeroc_fit(x=rr$x, t=rr$t, event=rr$event, obj = test)
jj <- timeroc_predict(cc, t = quantile(rr$t,probs = c(0.25, 0.5)))
plot(x = 1-jj[[1]][,2], y = jj[[1]][,1], type = 'l')
lines(x = 1-jj[[2]][,2], y = jj[[2]][,1], col = 'blue')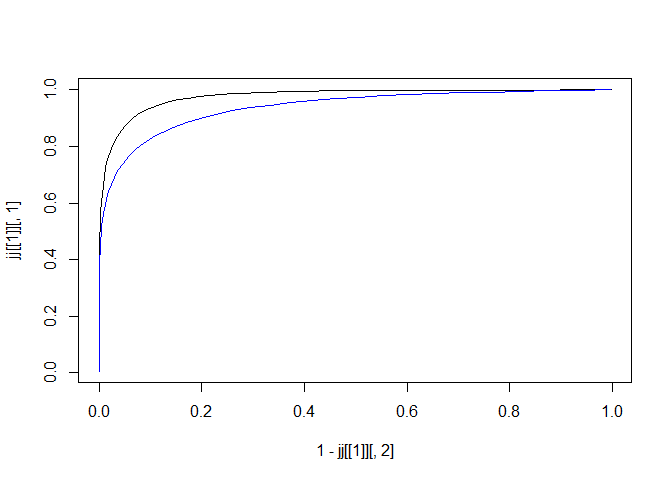
Fig.6. ROC curve at 25th & 50th quantile points of time-to-event
We can also specify the number of bootstrap process that we want if
confidence interval of the ROC curve need to be computed. The bootstrap
procedure can be achieved by supplying B = bootstrap value
into the parTimeROC::timeroc_predict() function.
library(parTimeROC)
# Copula model
test <- timeroc_obj(dist = 'gompertz-gompertz-copula', copula='clayton90',
params.t = c(shape=3,rate=1),
params.x = c(shape=1,rate=2),
params.copula=-5)
set.seed(23456)
rr <- rtimeroc(obj = test, censor.rate = 0.2, n=500)
cc <- timeroc_fit(x=rr$x, t=rr$t, event=rr$event, obj = test)
jj <- timeroc_predict(cc, t = quantile(rr$t,probs = c(0.25)), B = 500)
plot(x = 1-jj[[1]][,2], y = jj[[1]][,1], type = 'l')
lines(x = 1-jj[[1]][,4], y = jj[[1]][,3], col = 'red')
lines(x = 1-jj[[1]][,6], y = jj[[1]][,5], col = 'red')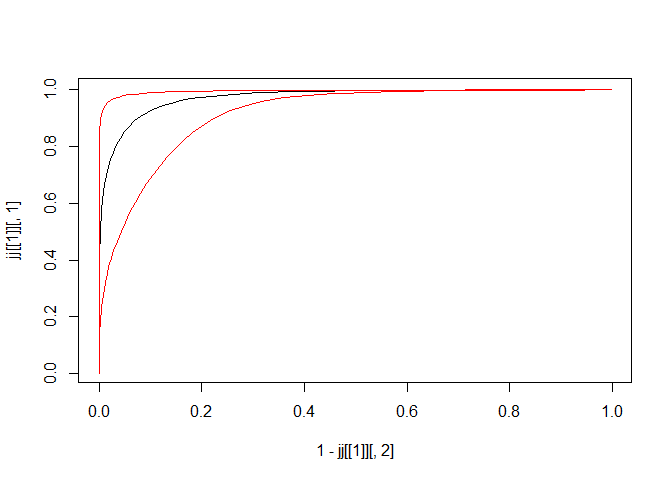
Fig.7. 95% boot confidence interval of ROC curve at 25th time-to-event
timeroc_aucFunction to compute the area under the ROC curve using the
parTimeROC::timeroc_auc() is also prepared for user
convenience.
test <- timeroc_obj('normal-weibull-copula', copula = 'clayton90')
print(test)
#> Model Assumptions: 90 Degrees Rotated Clayton Copula
#> X : Gaussian
#> Time-to-Event : Weibull
set.seed(23456)
rr <- rtimeroc(obj = test, censor.rate = 0.1, n=500,
params.t = c(shape=1, scale=5),
params.x = c(mean=5, sd=1),
params.copula=-2)
cc <- timeroc_fit(x=rr$x, t=rr$t, event=rr$event, obj = test)
jj <- timeroc_predict(cc, t = quantile(rr$t, probs = c(0.25,0.5,0.75)),
B = 500)
print(timeroc_auc(jj))
#> time assoc est.auc low.auc upp.auc
#> 1 1.671625 -1.889754 0.8871412 0.8360745 0.9251444
#> 2 3.822324 -1.889754 0.8204138 0.7650090 0.8657244
#> 3 7.396509 -1.889754 0.7725274 0.7156493 0.8204064These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.