
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.

The parafac4microbiome package enables R users with an
easy way to create Parallel Factor Analysis (PARAFAC) models for
longitudinal microbiome data.
processDataCube() can be used to process the microbiome
count data appropriately for a multi-way data array.parafac() allows the user to create a Parallel Factor
Analysis model of the multi-way data array.assessModelQuality() helps the user select the
appropriate number of components by randomly initializing many PARAFAC
models and inspecting various metrics of interest.assessModelStability() helps the user select the
appropriate number of components by bootstrapping or jack-knifing
samples and inspecting if the model outcome is similar.plotPARAFACmodel() helps visually inspect the PARAFAC
model.This package also comes with three example datasets.
Fujita2023: an in-vitro experiment of ocean inocula on
peptide medium, sampled every day for 110 days (doi:10.1186/s40168-023-01474-5).Shao2019: a large cohort dataset of vaginally and
caesarean-section born infants from London (doi:10.1038/s41586-019-1560-1).vanderPloeg2024: a small gingivitis intervention
dataset with response groups (doi:10.1101/2024.03.18.585469).A basic introduction to the package is given in
vignette("Introduction") and modelling the example datasets
are elaborated in their respective vignettes
vignette("Fujita2023"), vignette("Shao2019")
and vignette("vanderPloeg2024").
These vignettes and all function documentation can be found here.
The parafac4microbiome package can be installed from
CRAN using:
install.packages("parafac4microbiome")You can install the development version of
parafac4microbiome from GitHub with:
# install.packages("devtools")
devtools::install_github("GRvanderPloeg/parafac4microbiome")Please use the following citation when using this package:
library(parafac4microbiome)
set.seed(123)
# Process the data cube
processedFujita = processDataCube(Fujita2023,
sparsityThreshold=0.99,
CLR=TRUE,
centerMode=1,
scaleMode=2)
# Make a PARAFAC model
model = parafac(processedFujita$data, nfac=3, nstart=10, output="best")
# Sign flip components to make figure interpretable and comparable to the paper.
# This has no effect on the model or the fit.
model$Fac[[1]][,2] = -1 * model$Fac[[1]][,2] # sign flip mode 1 component 2
model$Fac[[2]][,1] = -1 * model$Fac[[2]][,1] # sign flip mode 2 component 1
model$Fac[[2]][,3] = -1 * model$Fac[[2]][,3] # sign flip mode 2 component 3
model$Fac[[3]] = -1 * model$Fac[[3]] # sign flip all of mode 3
# Plot the PARAFAC model using some metadata
plotPARAFACmodel(model$Fac, processedFujita,
numComponents = 3,
colourCols = c("", "Genus", ""),
legendTitles = c("", "Genus", ""),
xLabels = c("Replicate", "Feature index", "Time point"),
legendColNums = c(0,5,0),
arrangeModes = c(FALSE, TRUE, FALSE),
continuousModes = c(FALSE,FALSE,TRUE),
overallTitle = "Fujita PARAFAC model")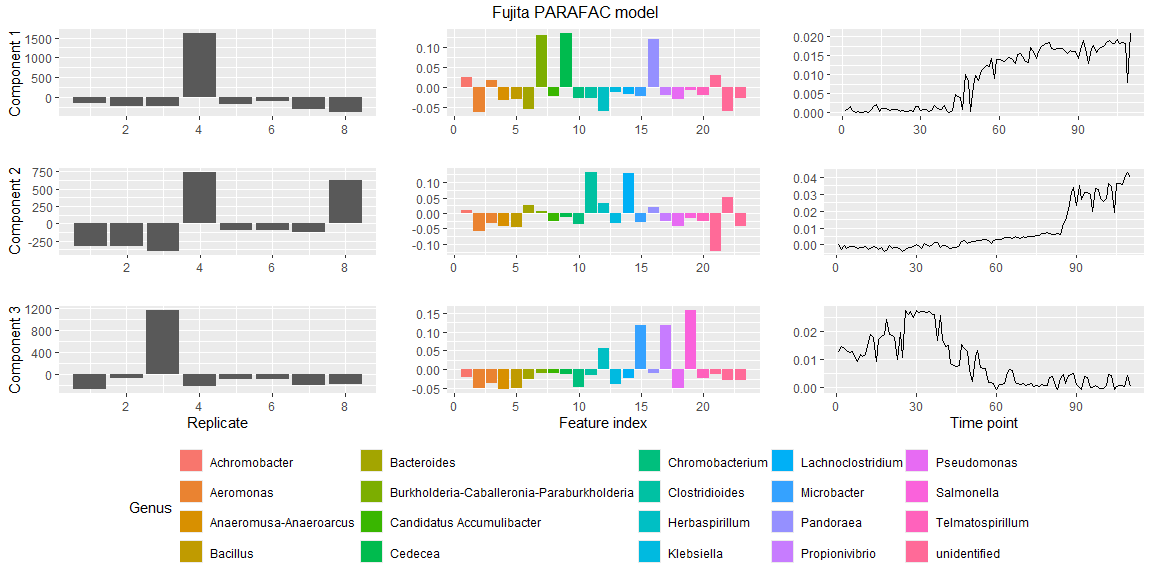
If you encounter an unexpected error or a clear bug, please file an issue with a minimal reproducible example here on Github. For questions or other types of feedback, feel free to send an email.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.