The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.

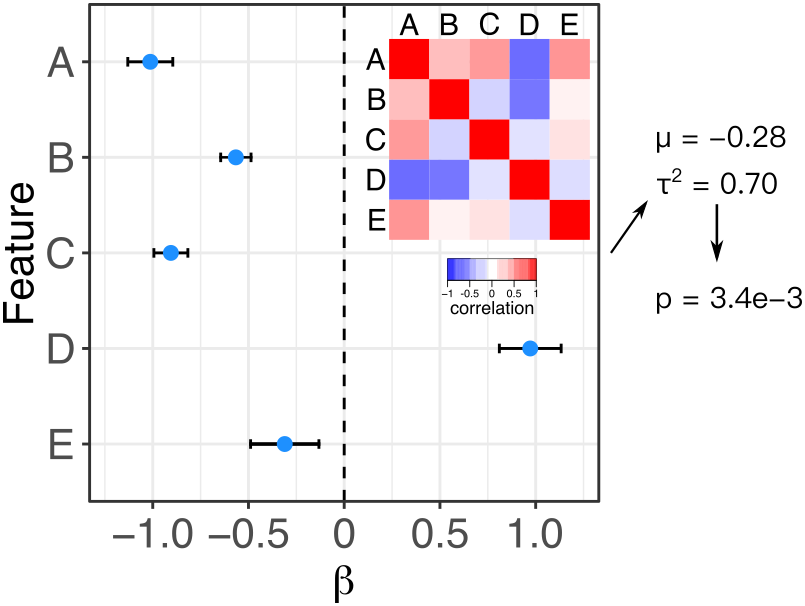
Meta-analysis is widely used to summarize estimated effects sizes across multiple statistical tests. Standard fixed and random effect meta-analysis methods assume that the estimated of the effect sizes are statistically independent. Here we relax this assumption and enable meta-analysis when the correlation matrix between effect size estimates is known. Fixed effect meta-analysis uses the method of Lin and Sullivan (2009), and random effects meta-analysis uses the method of Han, et al. 2016. An exentsion of the Lin-Sullivan method for finite sample size is described in Hoffman and Roussos (2025).
# Run fixed effects meta-analysis,
# accounting for correlation
LS( beta, stders, Sigma)
# Run fixed effects meta-analysis,
# accounting for correlation,
# and finite sample size using residual degrees of freedom
LS.empirical( beta, stders, Sigma, nu=rdf)
# Run random effects meta-analysis,
# accounting for correlation
RE2C( beta, stders, Sigma)devtools::install_github("DiseaseNeurogenomics/remaCor")These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.