
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.

scfetch is designed to accelerate users download and prepare single-cell datasets from public resources. It can be used to:
GEO/SRA, foramt fastq files to standard style that can be identified by 10x softwares (e.g. CellRanger).GEO/SRA, support downloading original 10x generated bam files (with custom tags) and normal bam files, and convert bam files to fastq files.GEO, PanglanDB and UCSC Cell Browser, load the downnloaded matrix to Seurat.Zeenodo, CELLxGENE and Human Cell Atlas.SeuratObject, AnnData, SingleCellExperiment, CellDataSet/cell_data_set and loom).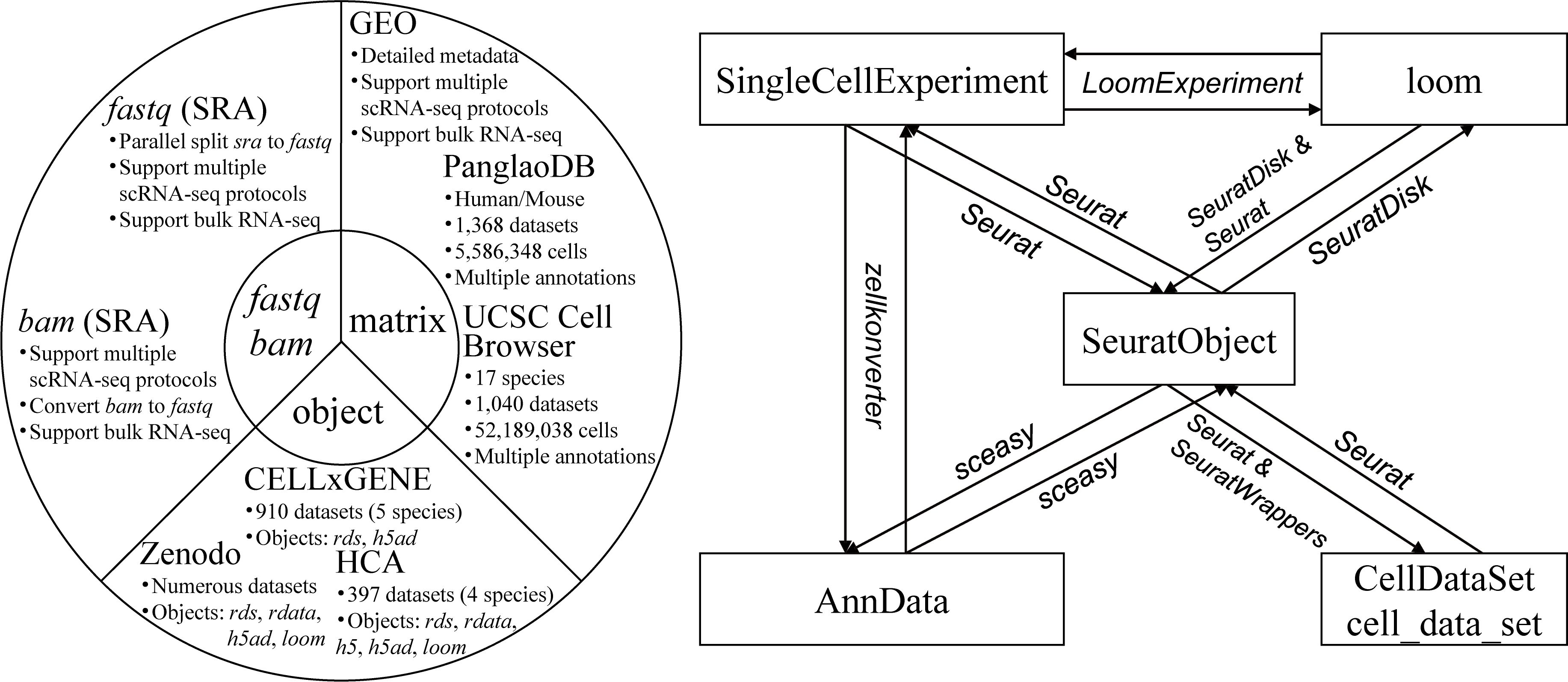
You can install the development version of scfetch from GitHub with:
For issues about installation, please refer INSTALL.md.
For data structures conversion, scfetch requires several python pcakages, you can install with:
Detailed usage is available in website.
| Type | Function | Usage |
|---|---|---|
| Download and format fastq | ExtractRun | Extract runs with GEO accession number or GSM number |
| DownloadSRA | Download sra files | |
| SplitSRA | Split sra files to fastq files and format to 10x standard style | |
| Download and convert bam | DownloadBam | Download bam (support 10x original bam) |
| Bam2Fastq | Convert bam files to fastq files | |
| Download matrix and load to Seurat | ExtractGEOMeta | Extract sample metadata from GEO |
| ParseGEO | Download matrix from GEO and load to Seurat | |
| ExtractPanglaoDBMeta | Extract sample metadata from PandlaoDB | |
| ExtractPanglaoDBComposition | Extract cell type composition of PanglaoDB datasets | |
| ParsePanglaoDB | Download matrix from PandlaoDB and load to Seurat | |
| ShowCBDatasets | Show all available datasets in UCSC Cell Browser | |
| ExtractCBDatasets | Extract UCSC Cell Browser datasets with attributes | |
| ExtractCBComposition | Extract cell type composition of UCSC Cell Browser datasets | |
| ParseCBDatasets | Download UCSC Cell Browser datasets and load to Seurat | |
| Download objects | ExtractZenodoMeta | Extract sample metadata from Zenodo with DOIs |
| ParseZenodo | Download rds/rdata/h5ad/loom from Zenodo with DOIs | |
| ShowCELLxGENEDatasets | Show all available datasets in CELLxGENE | |
| ExtractCELLxGENEMeta | Extract metadata of CELLxGENE datasets with attributes | |
| ParseCELLxGENE | Download rds/h5ad from CELLxGENE | |
| ShowHCAProjects | Show all available projects in Human Cell Atlas | |
| ExtractHCAMeta | Extract metadata of Human Cell Atlas projects with attributes | |
| ParseHCA | Download rds/rdata/h5/h5ad/loom from Human Cell Atlas | |
| Convert between different single-cell objects | ExportSeurat | Convert SeuratObject to AnnData, SingleCellExperiment, CellDataSet/cell_data_set and loom |
| ImportSeurat | Convert AnnData, SingleCellExperiment, CellDataSet/cell_data_set and loom to SeuratObject | |
| SCEAnnData | Convert between SingleCellExperiment and AnnData | |
| SCELoom | Convert between SingleCellExperiment and loom | |
| Summarize datasets based on attributes | StatDBAttribute | Summarize datasets in PandlaoDB, UCSC Cell Browser and CELLxGENE based on attributes |
For any question, feature request or bug report please write an email to songyb0519@gmail.com.
Please note that the scfetch project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.