The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.

The untb package provides R-centric functionality for
working with Hubbell’s unified neutral theory of biogeography and
biodiversity. A vignette is provided in the package. The canonical
reference is Hubbell 2001; to cite the package in publications please
use Hankin 2007.
You can install the released version of the untb package
from CRAN with:
# install.packages("untb") # uncomment this to install the package
library("untb")
set.seed(0)untb package in
useThe package has two main classes, count and
census. A count object is a named integer
vector, with names being species and entries being respective counts.
Thus:
x <- count(c(cats=9,pigs=3,dogs=2,rats=1,hogs=1,bats=1))
x
#> cats pigs dogs rats hogs bats
#> 9 3 2 1 1 1
summary(x)
#> Number of individuals: 17
#> Number of species: 6
#> Number of singletons: 3
#> Most abundant species: cats (9 individuals)
#> estimated theta: 2.861392Above, we see 9 cats, 3 pigs, and so on. Function
summary() gives further information. A census
object is an unordered factor with entries being the species of each
individual:
as.census(x)
#> [1] cats cats cats cats cats cats cats cats cats pigs pigs pigs dogs dogs rats
#> [16] hogs bats
#> Levels: cats pigs dogs rats hogs batsThe package includes example datasets:
data(sahfos)
summary(sahfos)
#> Number of individuals: 460182
#> Number of species: 54
#> Number of singletons: 10
#> Most abundant species: Echinodermata larvae (247200 individuals)
#> estimated theta: 4.649568We can give a visual summary of a dataset in two ways:
plot(sahfos)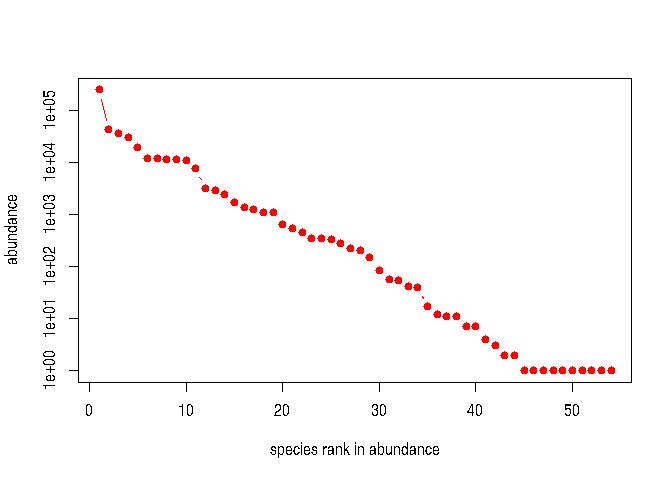
plot(preston(sahfos))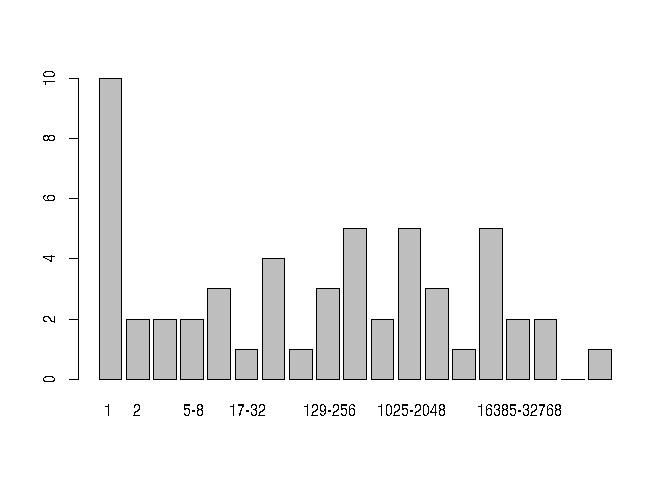
The package also includes the ability to generate random neutral assemblages:
summary(rand.neutral(1000,10))
#> Number of individuals: 1000
#> Number of species: 47
#> Number of singletons: 8
#> Most abundant species: 2 (330 individuals)
#> estimated theta: 10.09543
summary(rand.neutral(1000,10))
#> Number of individuals: 1000
#> Number of species: 52
#> Number of singletons: 16
#> Most abundant species: 4 (117 individuals)
#> estimated theta: 11.50422S. P. Hubbell 2001. “The Unified Neutral Theory of Biodiversity and Biogeography”, Princeton University Press.
R. K. S. Hankin, 2007. “Introducing untb, an R package for simulating ecological drift under the unified nuetral theory of biodiversity”, Journal of Statistical Software 22(12)
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.