The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
BayesMallowsSMC2 provides functions for performing sequential inference in the Bayesian Mallows model using the SMC2 algorithm (Sørensen et al. 2025).
You can install the development version of BayesMallowsSMC2 from GitHub with:
# install.packages("devtools")
devtools::install_github("osorensen/BayesMallowsSMC2")Here is a basic example using the included
complete_rankings dataset:
library(BayesMallowsSMC2)
# Fit the model with complete rankings
set.seed(123)
mod <- compute_sequentially(
complete_rankings,
hyperparameters = set_hyperparameters(n_items = 5),
smc_options = set_smc_options(n_particles = 100, n_particle_filters = 1)
)
# Show model info
mod
#> BayesMallowsSMC2 Model
#> ======================
#>
#> Number of particles: 100
#> Number of timepoints: 100
#> Number of items: 5
#> Number of clusters: 1
#>
#> Log marginal likelihood: -472.34
#> Final ESS: 63.55
#> Resampling events: 5/100We can visualize the posterior distributions of the parameters:
# Posterior distribution of alpha (dispersion parameter)
plot(mod, parameter = "alpha")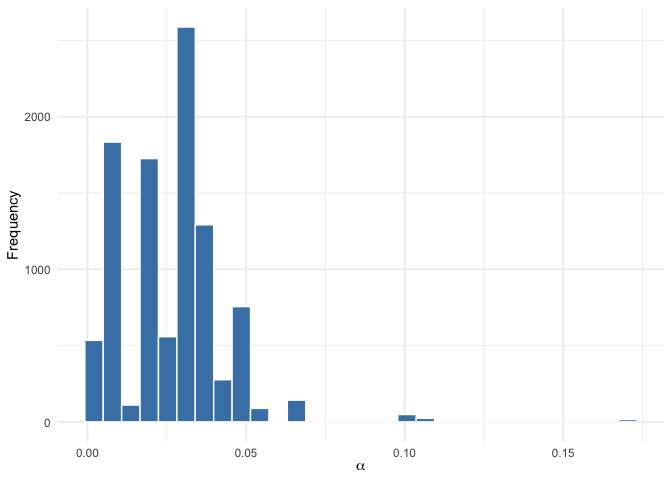
# Posterior distribution of rho (ranking positions)
plot(mod, parameter = "rho", items = c(1, 2, 3))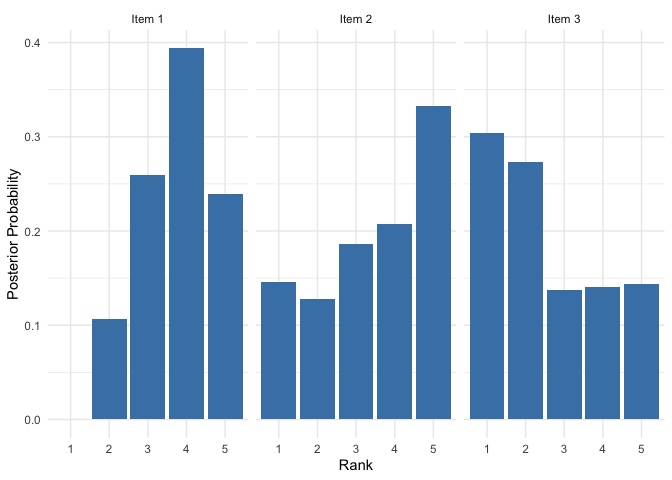
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.