The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
Cure dependent censoring regression models for long-term survival multivariate data.
You can install the development version of CureDepCens from GitHub with:
install.packages("devtools")
devtools::install_github("GabrielGrandemagne/CureDepCens")This is a basic example which shows you how to solve a common problem:
library(devtools)
#> Carregando pacotes exigidos: usethis
library(CureDepCens)
load_all()
#> ℹ Loading CureDepCens
Dogs_MimicData <- Dogs_MimicData
delta_t = ifelse(Dogs_MimicData$cens==1,1,0)
delta_c = ifelse(Dogs_MimicData$cens==2,1,0)
# MEP
fit <- cure_dep_censoring(formula = time ~ x1_cure + x2_cure | x_c1 + x_c2,
data = Dogs_MimicData,
delta_t = delta_t,
delta_c = delta_c,
ident = Dogs_MimicData$ident,
dist = "mep")
summary_cure(fit)
#>
#> MEP approach
#>
#> Name Estimate Std. Error CI INF CI SUP p-value
#> Alpha 2.034930 0.2005083 1.641933 2.427926 3.044e-26
#> Theta 0.7787554 0.4238412 0.000000 1.609484
#>
#> Coefficients Cure:
#>
#> Name Estimate Std. Error CI INF CI SUP p-value
#> Interc -0.6976047 0.1781988 -1.046874 -0.3483351 7.141e-33
#> x1_cur 0.514533 0.1703999 0.1805492 0.8485168 7.419e-18
#> x2_cur 0.2017428 0.08103922 0.04290593 0.3605797 0.001578
#>
#> Coefficients C:
#>
#> Name Estimate Std. Error CI INF CI SUP p-value
#> x_c1 0.03219111 0.1625781 -0.286462 0.3508442 0.1122
#> x_c2 -0.318467 0.1609394 -0.6339082 -0.003025754 4.682e-12
#>
#> ----------------------------------------------------------------------------------
#>
#> Information criteria:
#>
#> AIC BIC HQ
#> 510.9032 574.7666 536.194Dogs_MimicData is our simulated data frame. For more information check the documentation for stored datasets.
head(Dogs_MimicData)
#> u v t c time event int x1_cure
#> 1 0.56788087 0.83359383 0.4131564 0.3614745 0.3614745 0 1 0
#> 2 0.66013804 0.72909631 1.0968927 2.1033648 1.0968927 1 1 1
#> 3 0.06854872 0.63332194 Inf 1.6510975 1.6510975 0 1 1
#> 4 0.88345952 0.57152197 0.6522436 8.6456149 0.6522436 1 1 1
#> 5 0.45431855 0.92452776 0.9258282 0.5216269 0.5216269 0 1 1
#> 6 0.12120571 0.02350277 Inf 10.9070711 5.1121398 0 1 1
#> x2_cure x_c1 x_c2 cens ident
#> 1 0.5228382 1.0403070 0 2 1
#> 2 -0.4207129 0.1071675 1 1 2
#> 3 -1.1207319 -1.4042911 0 2 3
#> 4 1.1764416 -0.7740067 1 1 4
#> 5 0.3891404 0.4973770 1 2 5
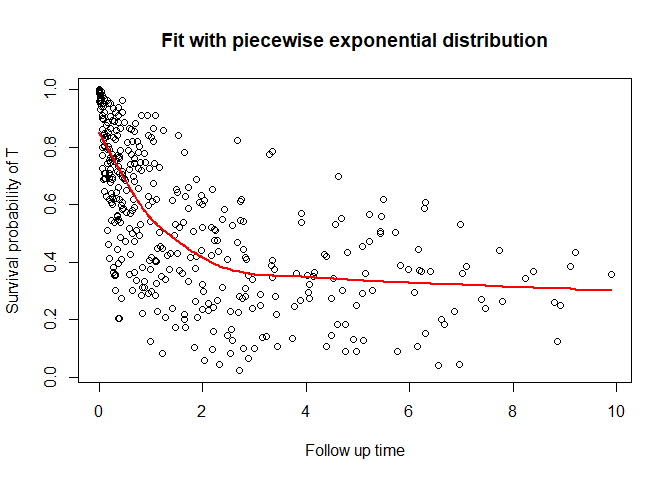
#> 6 0.5580893 -0.2278904 1 3 6You can also plot the survival function
plot_cure(fit, scenario = "t")
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.