The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
The EFAtools package provides functions to perform exploratory factor analysis (EFA) procedures and compare their solutions. The goal is to provide state-of-the-art factor retention methods and a high degree of flexibility in the EFA procedures. This way, implementations from R psych and SPSS can be compared. Moreover, functions for Schmid-Leiman transformation, and computation of omegas are provided. To speed up the analyses, some of the iterative procedures like principal axis factoring (PAF) are implemented in C++.
You can install the release version from CRAN with:
install.packages("EFAtools")You can install the development version from GitHub with:
install.packages("devtools")
devtools::install_github("mdsteiner/EFAtools")To also build the vignette when installing the development version, use:
install.packages("devtools")
devtools::install_github("mdsteiner/EFAtools", build_vignettes = TRUE)Here are a few examples on how to perform the analyses with the
different types and how to compare the results using the
COMPARE function. For more details, see the vignette by
running vignette("EFAtools", package = "EFAtools"). The
vignette provides a high-level introduction into the functionalities of
the package.
# load the package
library(EFAtools)
# Run multiple factor retention methods
N_FACTORS(test_models$baseline$cormat, N = 500)
#> Warning in N_FACTORS(test_models$baseline$cormat, N = 500): ! 'x' was a correlation matrix but CD needs raw data. Skipping CD.
#> ℹ The default implementation of EKC has changed compared to EFAtools version <= 0.5.0 to reflect the original version by Braeken and van Assen (2017). The previous version (which often yields different results from the original) is available with type = 'AM2019'. See details in the help page.
#>
#> ── Tests for the suitability of the data for factor analysis ───────────────────
#>
#> Bartlett's test of sphericity
#>
#> ✔ The Bartlett's test of sphericity was significant at an alpha level of .05.
#> These data are probably suitable for factor analysis.
#>
#> 𝜒²(153) = 2173.28, p < .001
#>
#> Kaiser-Meyer-Olkin criterion (KMO)
#>
#> ✔ The overall KMO value for your data is marvellous with 0.916.
#> These data are probably suitable for factor analysis.
#>
#> ── Number of factors suggested by the different factor retention criteria ──────
#>
#> • Comparison data: NA
#> • EKC (original implementation, type 'BvA2017'): 3
#> • Hull method with CAF: 3
#> • Hull method with CFI: 1
#> • Hull method with RMSEA: 1
#> • Parallel analysis with SMC: 3
#> • NEST: 3
# A type SPSS EFA to mimick the SPSS implementation with
# promax rotation
EFA_SPSS <- EFA(test_models$baseline$cormat, n_factors = 3, type = "SPSS",
rotation = "promax")
# look at solution
EFA_SPSS
#>
#> EFA performed with type = 'SPSS', method = 'PAF', and rotation = 'promax'.
#>
#> ── Rotated Loadings ────────────────────────────────────────────────────────────
#>
#> F1 F2 F3 h2 u2
#> V1 -.048 .035 .613 .367 .633
#> V2 -.001 .067 .482 .277 .723
#> V3 .060 .056 .453 .283 .717
#> V4 .101 -.009 .551 .378 .622
#> V5 .157 -.018 .438 .293 .707
#> V6 -.072 -.049 .704 .399 .601
#> V7 .001 .533 .093 .357 .643
#> V8 -.016 .581 .030 .349 .651
#> V9 .038 .550 -.001 .330 .670
#> V10 -.022 .674 -.071 .383 .617
#> V11 .015 .356 .232 .297 .703
#> V12 .020 .651 -.010 .432 .568
#> V13 .614 .086 -.067 .394 .606
#> V14 .548 -.068 .088 .322 .678
#> V15 .561 .128 -.070 .363 .637
#> V16 .555 -.050 .091 .344 .656
#> V17 .664 -.037 -.027 .390 .610
#> V18 .555 .004 .050 .350 .650
#>
#> ── Factor Intercorrelations ────────────────────────────────────────────────────
#>
#> F1 F2 F3
#> F1 1.000 0.617 0.648
#> F2 0.617 1.000 0.632
#> F3 0.648 0.632 1.000
#>
#> ── Variances Accounted for ─────────────────────────────────────────────────────
#>
#> F1 F2 F3
#> SS loadings 2.198 2.074 2.034
#> Prop Tot Var 0.122 0.115 0.113
#> Cum Prop Tot Var 0.122 0.237 0.350
#> Prop Comm Var 0.349 0.329 0.323
#> Cum Prop Comm Var 0.349 0.677 1.000
#>
#> ── Model Fit ───────────────────────────────────────────────────────────────────
#>
#> CAF: .50
#> df: 102
# A type psych EFA to mimick the psych::fa() implementation with
# promax rotation
EFA_psych <- EFA(test_models$baseline$cormat, n_factors = 3, type = "psych",
rotation = "promax")
# compare the type psych and type SPSS implementations
COMPARE(EFA_SPSS$rot_loadings, EFA_psych$rot_loadings,
x_labels = c("SPSS", "psych"))
#> Mean [min, max] absolute difference: 0.0090 [ 0.0001, 0.0245]
#> Median absolute difference: 0.0095
#> Max decimals where all numbers are equal: 0
#> Minimum number of decimals provided: 17
#>
#> F1 F2 F3
#> V1 0.0150 0.0142 -0.0195
#> V2 0.0109 0.0109 -0.0138
#> V3 0.0095 0.0103 -0.0119
#> V4 0.0118 0.0131 -0.0154
#> V5 0.0084 0.0105 -0.0109
#> V6 0.0183 0.0169 -0.0245
#> V7 -0.0026 -0.0017 0.0076
#> V8 -0.0043 -0.0035 0.0102
#> V9 -0.0055 -0.0040 0.0117
#> V10 -0.0075 -0.0066 0.0151
#> V11 0.0021 0.0029 0.0001
#> V12 -0.0064 -0.0050 0.0136
#> V13 -0.0109 -0.0019 0.0163
#> V14 -0.0049 0.0028 0.0070
#> V15 -0.0107 -0.0023 0.0161
#> V16 -0.0051 0.0028 0.0074
#> V17 -0.0096 -0.0001 0.0136
#> V18 -0.0066 0.0014 0.0098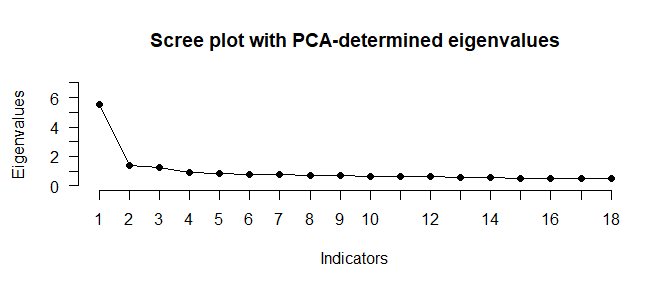
# Average solution across many different EFAs with oblique rotations
EFA_AV <- EFA_AVERAGE(test_models$baseline$cormat, n_factors = 3, N = 500,
method = c("PAF", "ML", "ULS"), rotation = "oblique",
show_progress = FALSE)
# look at solution
EFA_AV
#>
#> Averaging performed with averaging method mean (trim = 0) across 162 EFAs, varying the following settings: method, init_comm, criterion_type, start_method, rotation, k_promax, P_type, and varimax_type.
#>
#> The error rate is at 0%. Of the solutions that did not result in an error, 100% converged, 0% contained Heywood cases, and 100% were admissible.
#>
#>
#> ══ Indicator-to-Factor Correspondences ═════════════════════════════════════════
#>
#> For each cell, the proportion of solutions including the respective indicator-to-factor correspondence. A salience threshold of 0.3 was used to determine indicator-to-factor correspondences.
#>
#> F1 F2 F3
#> V1 .11 .00 1.00
#> V2 .11 .00 1.00
#> V3 .11 .00 .94
#> V4 .11 .00 1.00
#> V5 .11 .00 .94
#> V6 .11 .00 1.00
#> V7 .11 .94 .00
#> V8 .11 1.00 .00
#> V9 .11 .94 .00
#> V10 .11 1.00 .00
#> V11 .11 .89 .00
#> V12 .11 1.00 .00
#> V13 1.00 .00 .00
#> V14 1.00 .00 .00
#> V15 1.00 .00 .00
#> V16 1.00 .00 .00
#> V17 1.00 .00 .00
#> V18 1.00 .00 .00
#>
#>
#> ══ Loadings ════════════════════════════════════════════════════════════════════
#>
#> ── Mean ────────────────────────────────────────────────────────────────────────
#>
#> F1 F2 F3
#> V1 .025 .048 .576
#> V2 .060 .077 .451
#> V3 .115 .066 .425
#> V4 .157 .007 .518
#> V5 .198 -.002 .412
#> V6 .002 -.028 .658
#> V7 .074 .497 .102
#> V8 .056 .538 .046
#> V9 .100 .510 .018
#> V10 .048 .625 -.046
#> V11 .082 .336 .228
#> V12 .094 .606 .007
#> V13 .597 .083 -.047
#> V14 .531 -.056 .093
#> V15 .548 .122 -.049
#> V16 .540 -.041 .097
#> V17 .633 -.033 -.009
#> V18 .542 .009 .060
#>
#> ── Range ───────────────────────────────────────────────────────────────────────
#>
#> F1 F2 F3
#> V1 0.513 0.086 0.239
#> V2 0.431 0.093 0.186
#> V3 0.394 0.108 0.179
#> V4 0.415 0.110 0.214
#> V5 0.315 0.122 0.177
#> V6 0.514 0.104 0.267
#> V7 0.527 0.255 0.089
#> V8 0.520 0.275 0.078
#> V9 0.470 0.276 0.080
#> V10 0.533 0.313 0.097
#> V11 0.482 0.176 0.102
#> V12 0.548 0.324 0.103
#> V13 0.081 0.289 0.114
#> V14 0.063 0.220 0.117
#> V15 0.091 0.280 0.107
#> V16 0.072 0.230 0.122
#> V17 0.108 0.270 0.124
#> V18 0.081 0.246 0.118
#>
#>
#> ══ Factor Intercorrelations from Oblique Solutions ═════════════════════════════
#>
#> ── Mean ────────────────────────────────────────────────────────────────────────
#>
#> F1 F2 F3
#> F1 1.000 0.431 0.518
#> F2 0.431 1.000 0.454
#> F3 0.518 0.454 1.000
#>
#> ── Range ───────────────────────────────────────────────────────────────────────
#>
#> F1 F2 F3
#> F1 0.000 1.276 0.679
#> F2 1.276 0.000 1.316
#> F3 0.679 1.316 0.000
#>
#>
#> ══ Variances Accounted for ═════════════════════════════════════════════════════
#>
#> ── Mean ────────────────────────────────────────────────────────────────────────
#>
#> F1 F2 F3
#> SS loadings 2.443 1.929 1.904
#> Prop Tot Var 0.136 0.107 0.106
#> Prop Comm Var 0.389 0.307 0.303
#>
#> ── Range ───────────────────────────────────────────────────────────────────────
#>
#> F1 F2 F3
#> SS loadings 2.831 1.356 1.291
#> Prop Tot Var 0.157 0.075 0.072
#> Prop Comm Var 0.419 0.215 0.215
#>
#>
#> ══ Model Fit ═══════════════════════════════════════════════════════════════════
#>
#> M (SD) [Min; Max]
#> 𝜒²: 101.73 (34.62) [53.23; 125.98]
#> df: 102
#> p: .369 (.450) [.054; 1.000]
#> CFI: 1.00 (.00) [1.00; 1.00]
#> RMSEA: .01 (.01) [.00; .02]
#> AIC: -102.27 (34.62) [-150.77; -78.02]
#> BIC: -532.16 (34.62) [-580.66; -507.91]
#> CAF: .50 (.00) [.50; .50]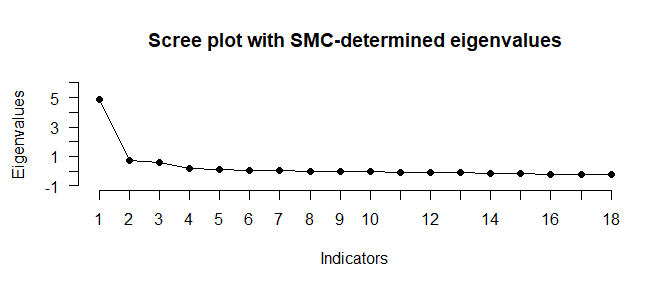
# Perform a Schmid-Leiman transformation
SL <- SL(EFA_psych)
# Based on a specific salience threshold for the loadings (here: .20):
factor_corres <- SL$sl[, c("F1", "F2", "F3")] >= .2
# Compute omegas from the Schmid-Leiman solution
OMEGA(SL, factor_corres = factor_corres)
#> Omega total, omega hierarchical, omega subscale, H index, explained common variance (ECV), and percent of uncontaminated correlations (PUC) for the general factor (top row) and omegas and H index for the group factors:
#>
#> tot hier sub H ECV PUC
#> g 0.883 0.750 0.122 0.845 0.668 0.706
#> F1 0.769 0.498 0.272 0.465
#> F2 0.764 0.494 0.270 0.473
#> F3 0.745 0.543 0.202 0.380If you use this package in your research, please acknowledge it by citing:
Steiner, M.D., & Grieder, S.G. (2020). EFAtools: An R package with fast and flexible implementations of exploratory factor analysis tools. Journal of Open Source Software, 5(53), 2521. https://doi.org/10.21105/joss.02521
If you want to contribute or report bugs, please open an issue on GitHub or email us at markus.d.steiner@gmail.com or silvia.grieder@gmail.com.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.