The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
The goal of InterpretMSSpectrum is to provides a set of R functions to annotate mass spectra from Electrospray-Ionization and Atmospheric-Pressure-Chemical-Ionization derived data in positive and negative ionization mode.
You can install the CRAN version of InterpretMSSpectrum using:
install.packages("InterpretMSSpectrum")or the development version from GitHub using:
# devtools is required to install from GitHub
# install.packages("devtools")
devtools::install_github("janlisec/InterpretMSSpectrum")In the simplest case InterpretMSSpectrum will provide an informed guess for the potential sum formula of an arbitrary mass spectrum.
# load APCI test data
apci_spectrum <- InterpretMSSpectrum::apci_spectrum
# find the most probable sum formula for the spectrum
# (will print to the console and open a new plot)
InterpretMSSpectrum::InterpretMSSpectrum(spec=apci_spectrum)
#>
#>
#> Top 8 of 13 remaining Formula combinations (ordered after mean score of combination)...
#> 73.75 56.52 26.27 21.26
#> 202.0893 "C8H16N1O3Si1" "C8H16N1O3Si1" "C8H16N1O3Si1" "C8H16N1O3Si1"
#> 246.1339 "C10H24N1O2Si2" "C11H24N1O1S1Si1" "C8H22N4O1Si2" NA
#> 274.1288 "C11H24N1O3Si2" "C12H24N1O2S1Si1" "C9H22N4O2Si2" NA
#> 348.1477 "C13H30N1O4Si3" "C14H30N1O3S1Si2" NA "C14H26N1O7Si1"
#> 364.1789 "C14H34N1O4Si3" "C15H34N1O3S1Si2" "C12H32N4O3Si3" "C15H30N1O7Si1"
#> 18.77 17.12 13.94 12.45
#> 202.0893 "C8H16N1O3Si1" "C6H14N4O2Si1" "C6H14N4O2Si1" NA
#> 246.1339 NA NA "C10H24N1O2Si2" NA
#> 274.1288 NA "C10H22N4O1S1Si1" "C9H22N4O2Si2" NA
#> 348.1477 "C9H33N2O4P1Si3" "C12H28N4O2S1Si2" NA "C13H34N1S1Si4"
#> 364.1789 "C10H37N2O4P1Si3" "C13H32N4O2S1Si2" "C10H30N7O2Si3" "C14H38N1S1Si4"
#>
#>
#> Total number of formulas per fragment before and after filtering...
#> mz initial score_cutoff PlausibleFormula TypicalLosses
#> 1 202.0893 30 4 2 2
#> 2 246.1339 69 28 4 4
#> 3 274.1288 114 43 4 4
#> 4 348.1477 339 118 10 10
#> 5 364.1789 402 166 13 13
#>
#>
#> Details of best candidate...
#> Formula Score Valid Mass
#> 1 C8H16N1O3Si1 75 Valid 202.0899456
#> 2 C10H24N1O2Si2 71 Valid 246.1345583
#> 3 C11H24N1O3Si2 44 Valid 274.1294729
#> 4 C13H30N1O4Si3 84 Valid 348.1482649
#> 5 C14H34N1O4Si3 79 Valid 364.1795650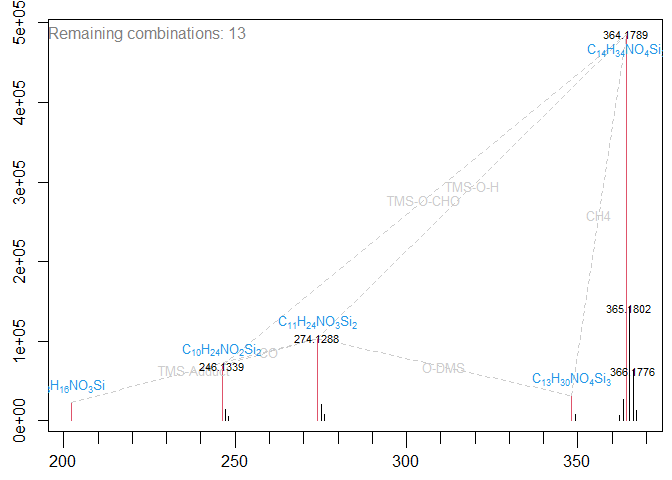
#>
#>
#> Time elapsed during individual processing steps...
#> Time differences in secs
#> FormulaGen ScoreFilt Plausible NeutralLoss PathEval Plot
#> 29.8296 0.0005 0.1235 0.1222 0.3729 0.0662The function can be tweaked with numerous parameters to limit the results, speed up calculations and more.
The other high level function of the package allows to predict the precursor of ESI spectra.
# load ESI test data
esi_spectrum <- InterpretMSSpectrum::esi_spectrum
# find the most likely precursor for the spectrum
(fmr <- InterpretMSSpectrum::findMAIN(spec=esi_spectrum))
#> Analyzed 102 neutral mass hypotheses (34 peaks * 3 adducts), kept 96
#> Selected m/z=263.0534 as [M+H]+ adduct of neutral mass 262.0461 with score 0.70.
#> mz int isogr iso charge adduct ppm label
#> 155 157.0501860 2.6500690604 NA NA NA <NA> NA <NA>
#> 229 193.0709159 29.2907409724 1 0 1 <NA> NA <NA>
#> 148 194.0747932 1.9549324775 1 1 1 <NA> NA <NA>
#> 117 196.9617186 1.0061243275 NA NA NA <NA> NA <NA>
#> 149 261.0377150 6.4563029839 NA NA NA <NA> NA <NA>
#> 233 263.0534072 100.0000000000 2 0 1 [M+H]+ NA [M+H]+
#> 187 264.0559143 6.1269254132 2 1 1 <NA> NA <NA>
#> 150 265.0579150 1.7051935769 2 2 1 <NA> NA <NA>
#> 140 278.0636869 3.3219943949 NA NA NA <NA> NA <NA>
#> 118 280.0799743 0.4884984457 NA NA NA <NA> NA <NA>
#> 119 283.0193302 2.4524382037 NA NA NA <NA> NA <NA>
#> 141 285.0353210 2.0414678564 NA NA NA [M+Na]+ 0.1012343615 [M+Na]+
#> 188 291.0490188 23.0130053610 NA NA NA <NA> NA <NA>
#> 121 308.0737825 1.4314882472 NA NA NA <NA> NA <NA>
#> 122 313.0305516 2.1848916230 NA NA NA <NA> NA <NA>
#> 123 359.0143852 1.0859216047 NA NA NA <NA> NA <NA>
#> 124 361.0297654 1.5472807922 NA NA NA <NA> NA <NA>
#> 125 389.0252820 1.6451046956 NA NA NA <NA> NA <NA>
#> 126 425.1053222 1.6750924050 NA NA NA <NA> NA <NA>
#> 127 453.1022657 3.4913206641 NA NA NA <NA> NA <NA>
#> 128 455.1181155 2.0655940307 NA NA NA <NA> NA <NA>
#> 129 483.1128831 2.0073900544 NA NA NA <NA> NA <NA>
#> 130 521.0697981 3.2406912531 NA NA NA <NA> NA <NA>
#> 154 523.0839382 9.7175989799 NA NA NA <NA> NA <NA>
#> 143 525.0990231 15.4786310623 3 0 1 [2M+H]+ 0.9805676686 [2M+H]+
#> 131 526.1044291 1.7618155343 3 1 1 <NA> NA <NA>
#> 144 551.0770120 10.4804797418 NA NA NA <NA> NA <NA>
#> 145 553.0947540 19.0018255376 4 0 1 <NA> NA <NA>
#> 146 554.0955402 3.1044267658 4 1 1 <NA> NA <NA>
#> 107 555.0985695 0.8381538242 4 2 1 <NA> NA <NA>
#> 108 561.0319968 0.8964040075 NA NA NA <NA> NA <NA>
#> 132 577.0014780 1.0216322103 NA NA NA <NA> NA <NA>
#> 147 581.0914486 10.4298634923 5 0 1 <NA> NA <NA>
#> 133 582.0933293 1.5373409114 5 1 1 <NA> NA <NA>
#> 113 617.1212331 0.5184300973 NA NA NA <NA> NA <NA>
#> 114 715.1447388 0.6167062300 NA NA NA <NA> NA <NA>
#> 135 783.1144323 2.8793012631 NA NA NA <NA> NA <NA>
#> 136 785.1256330 2.2229013611 NA NA NA <NA> NA <NA>
#> 137 811.1078594 2.8237031553 NA NA NA <NA> NA <NA>
#> 138 813.1263247 4.3365978759 NA NA NA <NA> NA <NA>
#> 115 815.1390540 1.6229672296 NA NA NA <NA> NA <NA>
#> 139 841.1211602 2.3651945193 NA NA NA <NA> NA <NA>
plot(fmr)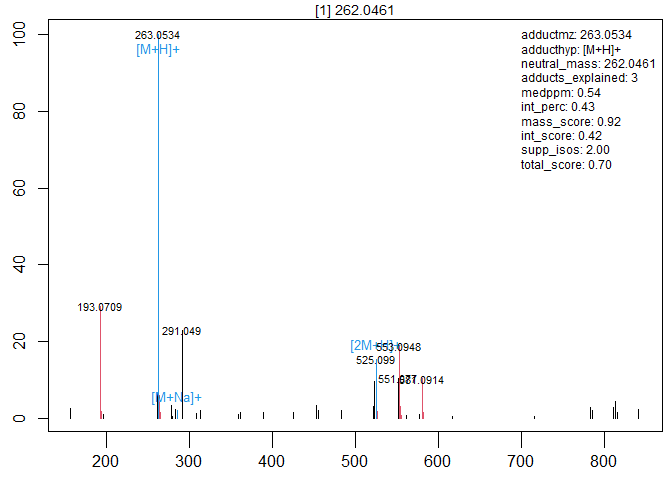
Also findMAIN provides multiple user options to use
individual adduct lists, thresholds and rule sets.
Finally, InterpretMSSpectrum provides a number of helper
functions, some of which are listed below.
# to count the chemical elements within a character vector of sum formulas
InterpretMSSpectrum::CountChemicalElements(x = "C6H12O6")
#> C H O
#> 6 12 6
sapply(c("C6H12O6", "CH3Cl"), InterpretMSSpectrum::CountChemicalElements, ele=c("C","H","O"))
#> C6H12O6 CH3Cl
#> C 6 1
#> H 12 3
#> O 6 0
# to get the exact mass for a sum formula
InterpretMSSpectrum::get_exactmass(c("C6H12O6", "Na", "H1"))
#> C6H12O6 Na H1
#> 180.063388200 22.989767700 1.007825035
# to check if one formula is contained in another (i.e. as fragment)
InterpretMSSpectrum:::is.subformula("H2O", "HCOOH")
#> O1C1
#> TRUEYou might read the publications on either APCI spectra processing which explains the idea of using the in source fragments for prediction of potential sum formulas or on ESI spectra processing which explains the strategy to infer the correct precursor of ESI mass spectra.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.