
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
The MFO package have been designed to calculate the Maximal Fat Oxidation (MFO), the exercise intensity that elicits MFO (Fatmax) and the SIN model to represent the fat oxidation kinetics. Three variables can be obtained from the SIN model: dilatation (d), symmetry (s) and traslation (t).Additionally, the package allows to calculate MFO and Fatmax of multiple subjects.
This is a basic example which shows you how to use the MFO package:
library(devtools)
install_github("JorgeDelro/MFO")
library(MFO)First, we have to load the data which consists in 3 databases: - basal_df: basal metabolism database. - MFO_df: MFO test database. - VO2max_df: results of a graded exercise test of which the VO2max of the subject is going to be extracted.
# Read dfs
data(list = c("basal_df", "MFO_df", "VO2max_df"), package = "MFO")
# Convert to data.frame
basal_df <- data.frame(basal_df)
MFO_df <- data.frame(MFO_df)
VO2max_df <- data.frame(VO2max_df)Then, we can used the function MFO
result_MFO <- MFO(step_time = 20,
db_MFO = MFO_df,
db_basal = basal_df,
db_graded = VO2max_df,
cv_var = "RER",
author = "Frayn",
VO2max = NULL)and the MFO can be plotted
print(result_MFO$MFO_plot)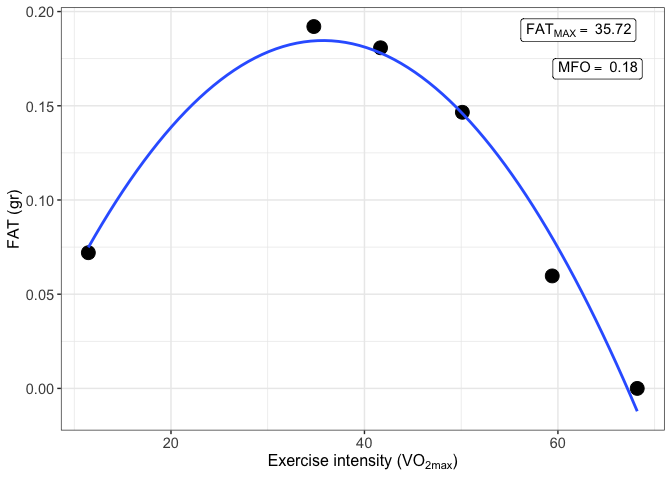
MFO kinetics are calculated using a database returned from MFO function called MFO_db.
result_MFO_kinetics <- MFO_kinetics(result_MFO$MFO_db)And again the function returns a plot with the results calculated
print(result_MFO_kinetics$MFO_kinetics_plot)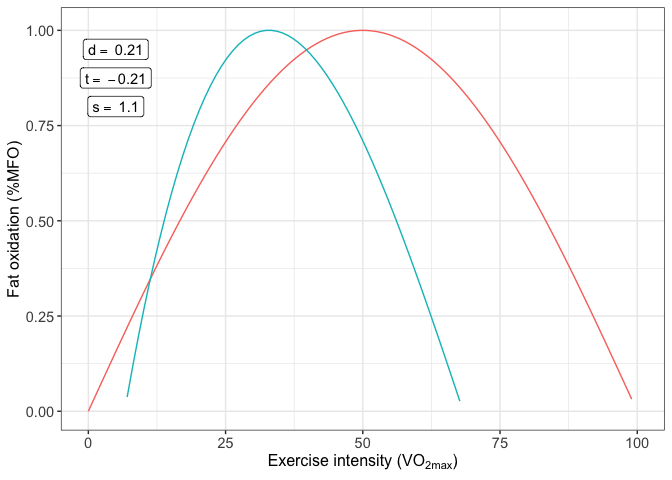
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.