
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.

Please make sure to read this at The NMsim website
where you can browse several vignettes with examples on specific
topics.
NMsim is an R package that can simulate Nonmem models
(using the NMsim() function) based on just a simulation
data set and a path to an estimation control stream. It will retrive and
combine output tables with input data once Nonmem has finished and
return the results to R.
The interface is “seamless” or fully integrated in R. Run a
simulation of the (estimated) model stored in “path/to/file.mod” using
the simulation input data set stored in the variable
data.sim this way:
simres <- NMsim(file.mod="/path/to/file.mod",
data=data.sim)You will quickly learn to do this on your own models, but if you can’t wait to see this working, you can do the following:
data.sim <- read.csv(system.file("examples/derived/dat_sim1.csv",package="NMsim"))
simres <- NMsim(file.mod=system.file("examples/nonmem/xgxr021.mod",package="NMsim"),
data=data.sim,
dir.sims=".")where dir.sims may be needed because the model in this
case may be in a read-only location.
Notice, file.mod could point to any working Nonmem model
as long as the provided simulation data set is sufficient to run it. We
are ready to plot:
library(ggplot2)
datl <- as.data.table(simres) |>
melt(measure.vars=cc(PRED,IPRED,Y))
ggplot(datl,aes(TIME,value,colour=variable))+
geom_line(data=function(x)x[variable!="Y"])+
geom_point(data=function(x)x[variable=="Y"])+
labs(x="Hours since first dose",y="Concentration (ng/mL)")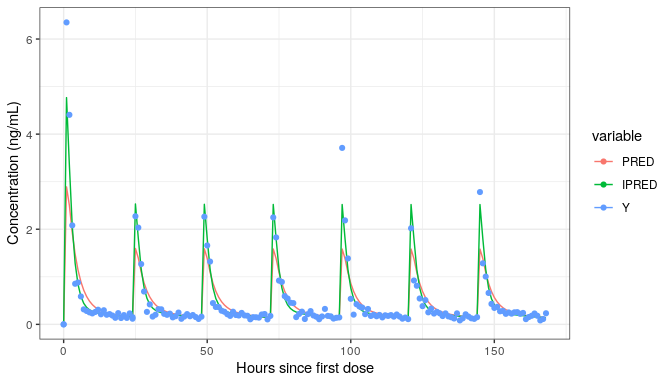
This example was a simulation of a multiple dose regimen with a
loading dose using a model estimated on single dose data. It is from the
first vignette NMsim-intro.html.
Go there next to get started with NMsim. The Examples
provides an overview of various tasks NMsim can help automating.
While NONMEM offers great flexibility for estimation of PK and PK/PD
models, many users find the simulation features in NONMEM insufficient
and turn to alternative software for simulation. This leads to
additional work of model reimplementation, with risk of the simulation
model deviating from the estimated model, due to bugs in the
reimplementation. For a wide range of model types, the limitation is not
in NONMEM’s ability to perform such simulations, but rather in the lack
of a simple user-interface to obtain the simulations. NMsim
provides such an interface as an R package, allowing the modeler to
simulate models as soon as an estimate is available.
The goal for NMsim is to automate the NONMEM simulation
workflow and provide a simple, flexible, and powerful R interface. This
allows for automation of most simulation-based analyses.
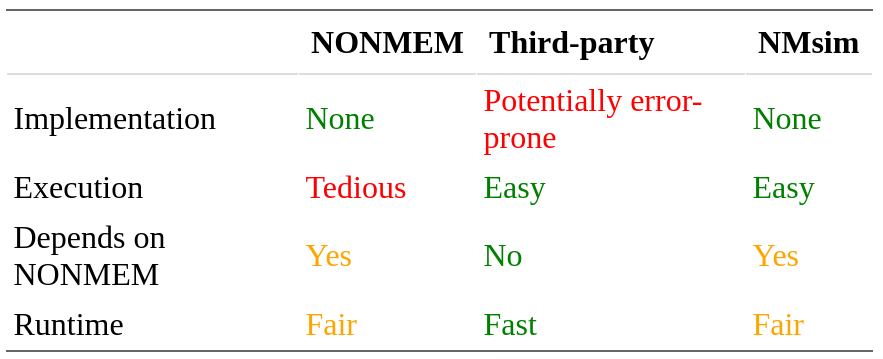
NMsim has a flexible way to define simulation methods.
The following methods are currently provided:
method.sim=NMsim_default)method.sim=NMsim_EBE)method.sim=NMsim_VarCov and
method.sim=NMsim_NWPRI)method.sim=NMsim_asis)In addition, NMsim provides other features to further
modify the simulation control stream
typical=TRUE)inits argument)modify
argument)To learn how to run these simulations on your Nonmem models, get
started with NMsim-intro.html.
It is really easy.
In addition, NMsim can simulate multiple models at a
time. E.g., if a bootstrap run of a model is available, NMsim can run
the simulation with each of the bootstrap models and collect all the
results in one dataset. This provides a robust and easy way to simulate
a Nonmem model with uncertainty.
You can also write your own methods, if you have some other
Nonmem-based simulation (or other job) you want to automate using
NMsim.
Many features are available. Prominent ones are:
NMreadSim() to read
previously performed simulation results.reuse.results=TRUE.SUBPROBLEMS feature
avaible through the subproblems argumenttransform
argument.If residual variability is not implemented in the simulated model,
NMsim provides a way (addResVar()) to add
residual variability in R after the simulation has been run.
NMsim does not simulate, translate or otherwise
interpret a NONMEM model. Instead, it automates the NONMEM simulation
workflow (including execution of NONMEM) and wraps it all into one R
function. Provided with a path to a NONMEM control stream and a
data.frame to simulate, NMsim will do the following:
file.mod ($INPUT and $DATA
matching the saved simulation data set; $SIMULATE instead
of $ESTIMATION and $COVARIANCE)file.ext)NMsim can call NONMEM directly or via PSN.
If NMsim is run on a system where NONMEM cannot be
executed, NMsim can still prepare the simulation control
stream and datafile.
NMsim is in itself a relatively small R package. It
makes extensive use of functionality to handle NONMEM data and control
streams provided by the R package NMdata.
The methods currently provided by NMsim will work with
(many or most) Pop PK models and most continuous-scale PD models.
Methods are currently not provided for for time-to-event models. Also,
depending on the coding of the models, other censored data models may
not work out of the box, because the model may not have a single
variable (in Nonmem) that simulates the wanted information for all data
rows, as their interpretation may depend on other values.
The input data set must contain whatever variables are needed by the
Nonmem model. A common issue is if the Nonmem model uses a covariate
that is not in the simulation input data set. NMdata’s NMcheckData
is a good help identifying input data issues before running Nonmem - and
when Nonmem acts unexpectedly.
Nonmem may not be the fastest simulator out there. But actually most
often, the reason Nonmem is slow at providing a simulation result is
that it takes a long time writing the $TABLE files (yes,
that can account for 90% or more of the time Nonmem spends).
NMsim provides a simple way to get around this. The
argument text.table can be used to define only the columns
needed in the simulation output (which may be as little as
PRED, IPRED, and a couple more - remember the
input data is merged back automatically). As a result,
NMsim may still be slower than a re-implementation in a
different framework. But it’s extremely easy to do.
NMsim is dependent on running Nonmem. Often, that will mean Nonmem
must be available on the same system as the one running R. However, if
Nonmem is run on a separate system through qsub or in
another way initiates Nonmem on another system, that will work too. Then
however, only if R can read the file system where Nonmem writes the
results, it can retrieve the results.
NMsim does not need PSN but can use it. However, not all features are
available with PSN, so for some features you will have to specify the
path to the Nonmem executable (say
path.nonmem=/path/to/nmfe75 or any Nonmem executable you
want to use). Specifically of the simulation types currently available,
simulation of known subjects is not possible using PSN (but works if a
Nonmem executable is provided).
If PSN is used, NMsim uses PSN’s execute to
run models. In addition, NMsim by default uses PSN’s
update_inits to update initial values in control streams,
if PSN is available. NMsim does also include its own simple
function to do this if PSN is not available.
NMsim reliable?Importantly, NMsim does not (at least not by default)
modify, translate or simulate the model itself. It does modify control
stream sections $INPUT, $DATA,
$ESTIMATION, $SIMULATION, $THETA,
$OMEGA, $SIGMA, $TABLE as needed.
The fact that NMsim allows for skipping the
re-implementation but just uses Nonmem to simulate the Nonmem model as
is, eliminates the risk of discrepancies between the estimated model and
the simulated model.
The produced control stream is saved together with simulation data
set open for manual inspection and can obviously be run with Nonmem
independently of NMsim.
NMsim includes functions (NMcreateDoses and
addEVID2) to very easily create simulation data sets. While
one certainly does not need to use these functions to use
NMsim, they do add to the package providing a framework
that enables a complete simulation workflow in only 5-15 simple lines of
R code.
There are several other packages out there that can do this, and
NMsim may not be your best choice if this feature is all
you are looking for. However, running Nonmem using the
NMexec() function provided by NMsim has one
important advantage in that it saves the input data together with the
Nonmem control streams. This ensures that output data can be merged with
input data as it went into the model, even if the input data file should
be modified or lost.
NMexec will submit model runs to a cluster by default.
This can be switched off for running Nonmem locally. Please notice the
jobs are submitted to a cluster in a very specific way using
PSN. If your setup is different, this is for now not
supported. Please use NMexec(sge=FALSE) in that case (which
may not be desirable). Notice that simulations are not done on a cluster
by default so you may still be able to use NMsim.
NMsim is on CRAN, MPN and github:
## From CRAN/MPN repositories
install.packages("NMsim")
## From github
library(remotes)
install_github("NMautoverse/NMsim")These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.