
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.

PFW is an R package designed for easy filtering,
preparation, and management of data from Project FeederWatch. Project
FeederWatch is a community-driven project initiated in the 1980s and run
by the Cornell Lab of
Ornithology and Birds
Canada that compiles bird observations from thousands of “backyards,
nature centers, community areas, and other locales” across North
America. Project FeederWatch data is easy to access, but can often be
tricky to work with; PFW serves to simplify and streamline
the use of this data. Included in PFW are tools for
taxonomic rollup, filtering by survey characteristics (species, state,
etc.), merging in site metadata, and zerofilling for presence/absence
modeling.
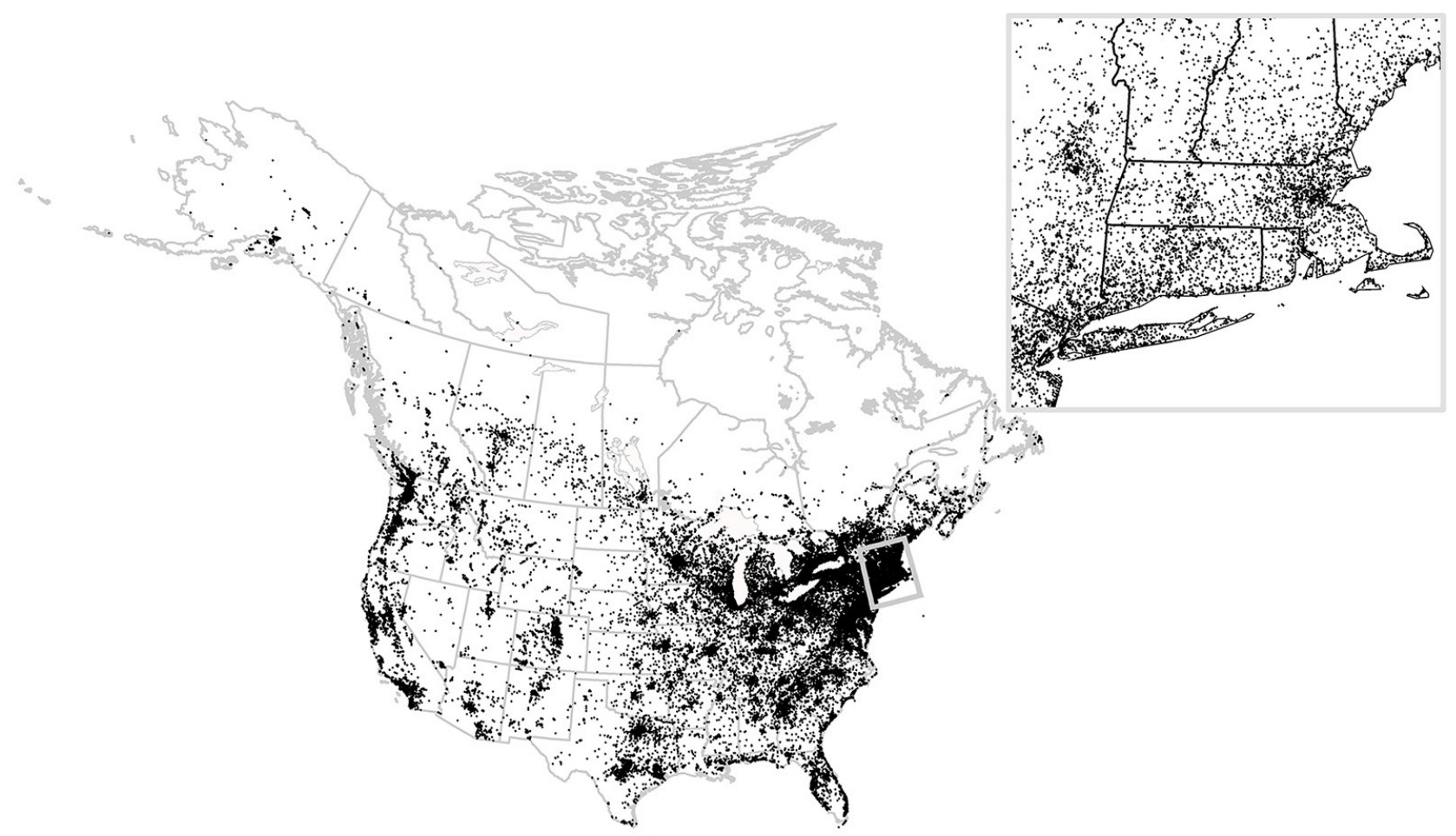
Figure from Bonter & Greig (2021), licensed under CC BY 4.0.
PFW can be installed using:
install.packages("PFW", repos = c('https://ropensci.r-universe.dev', 'https://cloud.r-project.org'))Background and details on using PFW to filter and
process Project FeederWatch data are outlined in the vignette.
To cite PFW in publications, use:
Maron, M. W. (2025). PFW: Filtering and Processing Data from Project FeederWatch.
R package version 0.1.0. https://github.com/ropensci/PFW
You can also run
citation("PFW")in R when PFW is loaded.
This is a simple example which shows you how to do basic Project FeederWatch importing, filtering, zerofilling, and site data attachment. Here, we’ll load the example dataset and filter it for Song Sparrow, Dark-eyed Junco, and Spotted Towhee from Washington and Oregon between 2022 and 2024 in November-February. Then, we’ll zerofill that data and attach our site metadata:
library(PFW)
# Load in the included example dataset
data <- pfw_example # If you were using your own selection of PFW data,
# this would be pfw_import() instead.
# pfw_import() creates and defaults to "/data-raw" in a local directory,
# but will accept a different filepath.
# Create a list of study species
species_list <- c("Song Sparrow", "Dark-eyed Junco", "Spotted Towhee")
# Create a list of study regions
region_list <- c("Washington", "Oregon")
# Filter data by species and region, from 2022–2024 during November-February
data_filtered <- pfw_filter(data,
region = region_list,
species = species_list,
year = 2022:2024,
month = 11:2, # pfw_date(), which is called within pfw_filter(),
# will appropriately wrap this around the end of the year.
valid = TRUE, # TRUE by default
rollup = TRUE # TRUE by default
)
# Output:
# 2 regions successfully filtered.
# Date filtering complete.
# Species roll-up complete. 36 ambiguous records removed.
# 3 species successfully filtered.
# Filtering complete. 23538 records remaining.
# View the filters that were applied
pfw_attr(data_filtered)
# Output:
# Filters applied to this dataset:
#
# - Filter type: region
# Values: Washington, Oregon
#
# - Filter type: date
# year : 2022, 2023, 2024
# month : 11, 12, 1, 2
#
# - Filter type: rollup
# Values: TRUE
#
# - Filter type: valid
# Values: TRUE
#
# - Filter type: species
# Values: song sparrow, dark-eyed junco, spotted towhee
# Zerofill missing species/survey instance combos
data_zf <- pfw_zerofill(data_filtered)
# Attach site description metadata
# Replace "path/sitedata.csv" with the actual path to the downloaded file
data_full <- pfw_sitedata(data_zf, path = "path/sitedata.csv")
# If the file does not exist at that path, pfw_sitedata will
# download it there from the Project FeederWatch website.
# Alternatively, you can manually download the site description file from:
# https://feederwatch.org/explore/raw-dataset-requests/Have feedback? Please submit any bugs, typos, or improvements as an
issue or as a pull request! Your
insights help improve PFW.
Please note that PFW is released with a Contributor
Code of Conduct. By contributing to this project, you agree to abide
by its terms.
PFW was built from code originally developed for Project
FeederWatch data preparation in Maron et al. (2025). While the function
scripts in this package were created specifically for PFW, the code they
are based on benefited greatly from examples and code snippets provided
by Emma Greig, who passed away prior to the package’s creation.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.