
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.

Chest pain? Calculate your cardiovascular risk score.
The goal of RiskScorescvd R package is to calculate the most commonly used cardiovascular risk scores. Original research publication can be found here: https://openheart.bmj.com/content/11/2/e002755
We have developed nine of the most commonly used risk scores with a dependency (ASCVD [PooledCohort]) making the following available:
NOTE: Troponin I values should be used. Additional functions for Troponin T are under development
You can install from CRAN with:
# Install from CRAN
install.packages("RiskScorescvd")You can install the development version of RiskScorescvd from GitHub with:
# install.packages("devtools")
devtools::install_github("dvicencio/RiskScorescvd")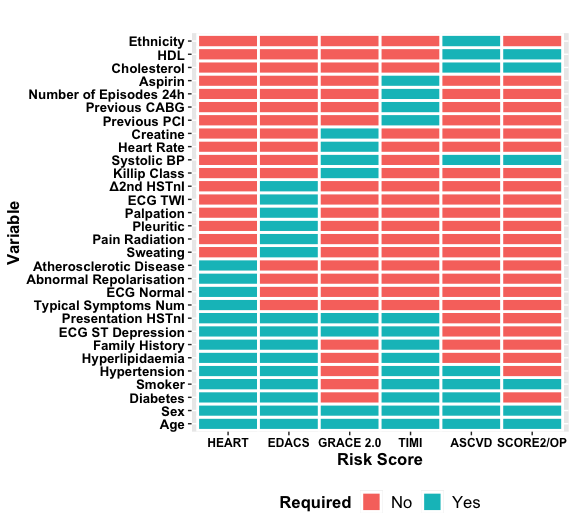
This is a basic example of how the data set should look to calculate all risk scores available in the package:
library(RiskScorescvd)
#> Loading required package: PooledCohort
# Create a data frame or list with the necessary variables
# Set the number of rows
num_rows <- 100
# Create a large dataset with 100 rows
cohort_xx <- data.frame(
typical_symptoms.num = as.numeric(sample(0:6, num_rows, replace = TRUE)),
ecg.normal = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
abn.repolarisation = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
ecg.st.depression = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
Age = as.numeric(sample(30:80, num_rows, replace = TRUE)),
diabetes = sample(c(1, 0), num_rows, replace = TRUE),
smoker = sample(c(1, 0), num_rows, replace = TRUE),
hypertension = sample(c(1, 0), num_rows, replace = TRUE),
hyperlipidaemia = sample(c(1, 0), num_rows, replace = TRUE),
family.history = sample(c(1, 0), num_rows, replace = TRUE),
atherosclerotic.disease = sample(c(1, 0), num_rows, replace = TRUE),
presentation_hstni = as.numeric(sample(10:100, num_rows, replace = TRUE)),
Gender = sample(c("male", "female"), num_rows, replace = TRUE),
sweating = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
pain.radiation = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
pleuritic = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
palpation = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
ecg.twi = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
second_hstni = as.numeric(sample(1:200, num_rows, replace = TRUE)),
killip.class = as.numeric(sample(1:4, num_rows, replace = TRUE)),
heart.rate = as.numeric(sample(0:300, num_rows, replace = TRUE)),
systolic.bp = as.numeric(sample(40:300, num_rows, replace = TRUE)),
aspirin = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
creat = as.numeric(sample(0:4, num_rows, replace = TRUE)),
number.of.episodes.24h = as.numeric(sample(0:20, num_rows, replace = TRUE)),
previous.pci = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
cardiac.arrest = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
previous.cabg = as.numeric(sample(c(0, 1), num_rows, replace = TRUE)),
total.chol = as.numeric(sample(5:100, num_rows, replace = TRUE)),
total.hdl = as.numeric(sample(2:5, num_rows, replace = TRUE)),
Ethnicity = sample(c("white", "black", "asian", "other"), num_rows, replace = TRUE),
eGFR = as.numeric(sample(15:120, num_rows, replace = TRUE)),
ACR = as.numeric(sample(5:1500, num_rows, replace = TRUE)),
trace = sample(c("trace", "1+", "2+", "3+", "4+"), num_rows, replace = TRUE)
)
str(cohort_xx)
#> 'data.frame': 100 obs. of 34 variables:
#> $ typical_symptoms.num : num 1 1 0 2 4 6 6 2 4 4 ...
#> $ ecg.normal : num 0 1 1 0 0 0 0 1 1 0 ...
#> $ abn.repolarisation : num 1 0 1 1 0 0 0 1 0 1 ...
#> $ ecg.st.depression : num 0 1 0 1 1 0 1 0 1 0 ...
#> $ Age : num 61 75 60 44 47 46 71 39 56 50 ...
#> $ diabetes : num 1 1 1 1 0 1 0 0 1 1 ...
#> $ smoker : num 0 1 0 1 0 0 1 1 1 1 ...
#> $ hypertension : num 1 1 1 1 1 0 1 0 0 0 ...
#> $ hyperlipidaemia : num 1 0 1 0 1 0 0 1 1 0 ...
#> $ family.history : num 1 0 0 0 1 1 0 1 0 1 ...
#> $ atherosclerotic.disease: num 1 1 1 0 1 0 0 1 1 0 ...
#> $ presentation_hstni : num 19 75 96 76 82 35 36 20 54 27 ...
#> $ Gender : chr "male" "female" "male" "female" ...
#> $ sweating : num 1 0 0 1 1 0 1 0 1 0 ...
#> $ pain.radiation : num 1 1 1 1 1 1 1 1 0 1 ...
#> $ pleuritic : num 1 1 0 0 0 1 1 1 0 0 ...
#> $ palpation : num 1 0 1 0 0 0 0 0 0 1 ...
#> $ ecg.twi : num 0 1 1 0 0 1 1 0 0 1 ...
#> $ second_hstni : num 157 131 58 45 174 23 178 189 20 78 ...
#> $ killip.class : num 3 4 4 3 2 3 2 2 1 4 ...
#> $ heart.rate : num 227 273 122 169 147 291 267 266 198 148 ...
#> $ systolic.bp : num 41 218 153 118 282 92 191 293 101 267 ...
#> $ aspirin : num 1 0 0 1 0 0 1 0 1 1 ...
#> $ creat : num 3 3 2 3 3 0 1 2 4 3 ...
#> $ number.of.episodes.24h : num 11 12 19 18 12 17 10 16 4 8 ...
#> $ previous.pci : num 0 0 0 1 0 1 0 1 1 0 ...
#> $ cardiac.arrest : num 0 1 0 0 1 1 0 0 1 1 ...
#> $ previous.cabg : num 0 1 1 0 0 0 0 1 0 0 ...
#> $ total.chol : num 97 89 52 93 29 56 58 99 91 40 ...
#> $ total.hdl : num 3 3 4 3 4 5 5 4 5 2 ...
#> $ Ethnicity : chr "other" "white" "white" "white" ...
#> $ eGFR : num 77 69 110 81 22 76 68 58 43 65 ...
#> $ ACR : num 782 1161 1005 595 840 ...
#> $ trace : chr "trace" "4+" "4+" "trace" ...This is a basic example of how to calculate all risk scores available in the package and create a new data set with 12 new variables of the calculated and classified risk scores:
# Call the function with the cohort_xx to calculate all risk scores available in the package
new_data_frame <- calc_scores(data = cohort_xx)
# Select columns created after calculation
All_scores <- new_data_frame %>% select(HEART_score, HEART_strat, EDACS_score, EDACS_strat, GRACE_score, GRACE_strat, TIMI_score, TIMI_strat, SCORE2_score, SCORE2_strat, ASCVD_score, ASCVD_strat)
# Observe the results
head(All_scores)
#> # A tibble: 6 × 12
#> # Rowwise:
#> HEART_score HEART_strat EDACS_score EDACS_strat GRACE_score GRACE_strat
#> <dbl> <ord> <dbl> <ord> <dbl> <ord>
#> 1 4 Moderate risk 14 Not low risk 150 High risk
#> 2 8 High risk 15 Not low risk 181 High risk
#> 3 5 Moderate risk 13 Not low risk 132 High risk
#> 4 7 High risk 14 Not low risk 131 High risk
#> 5 8 High risk 16 Not low risk 88 Low risk
#> 6 6 Moderate risk 5 Not low risk 132 High risk
#> # ℹ 6 more variables: TIMI_score <dbl>, TIMI_strat <ord>, SCORE2_score <dbl>,
#> # SCORE2_strat <ord>, ASCVD_score <dbl>, ASCVD_strat <ord>
# Create a summary of them to obtain an initial idea of distribution
summary(All_scores)
#> HEART_score HEART_strat EDACS_score EDACS_strat
#> Min. : 2.00 Low risk : 6 Min. :-2.00 Low risk : 1
#> 1st Qu.: 5.00 Moderate risk:54 1st Qu.: 5.00 Not low risk:99
#> Median : 6.00 High risk :40 Median :10.00
#> Mean : 6.06 Mean :10.78
#> 3rd Qu.: 7.00 3rd Qu.:16.00
#> Max. :10.00 Max. :30.00
#> GRACE_score GRACE_strat TIMI_score TIMI_strat
#> Min. : 29.00 Low risk :30 Min. :2.00 Very low risk: 0
#> 1st Qu.: 85.75 Moderate risk:34 1st Qu.:4.00 Low risk : 6
#> Median :107.00 High risk :36 Median :4.00 Moderate risk:49
#> Mean :108.53 Mean :4.44 High risk :45
#> 3rd Qu.:132.00 3rd Qu.:5.00
#> Max. :205.00 Max. :7.00
#> SCORE2_score SCORE2_strat ASCVD_score ASCVD_strat
#> Min. : 0.00 Very low risk: 0 Min. :0.0000 Very low risk: 7
#> 1st Qu.: 55.00 Low risk : 6 1st Qu.:0.1900 Low risk : 2
#> Median :100.00 Moderate risk: 6 Median :0.4650 Moderate risk:17
#> Mean : 75.87 High risk :88 Mean :0.5027 High risk :74
#> 3rd Qu.:100.00 3rd Qu.:0.8300
#> Max. :100.00 Max. :1.0000These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.