The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
To install from CRAN:
install.packages("Rtsne") # Install Rtsne package from CRANTo install the latest version from the github repository, use:
if(!require(devtools)) install.packages("devtools") # If not already installed
devtools::install_github("jkrijthe/Rtsne")After installing the package, use the following code to run a simple example (to install, see below).
library(Rtsne) # Load package
iris_unique <- unique(iris) # Remove duplicates
set.seed(42) # Sets seed for reproducibility
tsne_out <- Rtsne(as.matrix(iris_unique[,1:4])) # Run TSNE
plot(tsne_out$Y,col=iris_unique$Species,asp=1) # Plot the result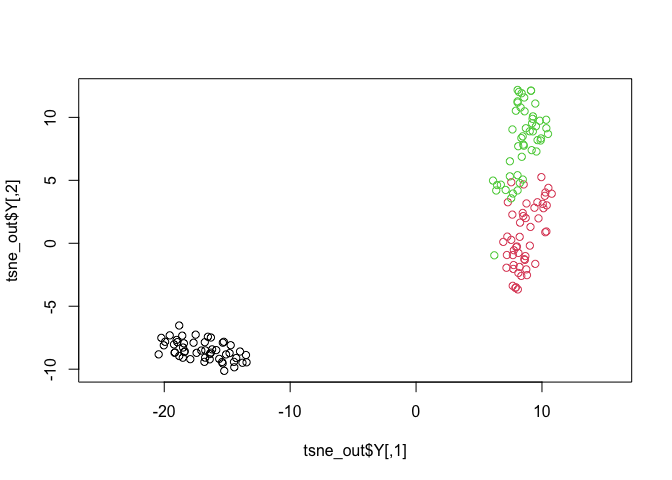
This R package offers a wrapper around the Barnes-Hut TSNE C++ implementation of [2] [3]. Changes were made to the original code to allow it to function as an R package and to add additional functionality and speed improvements.
[1] L.J.P. van der Maaten and G.E. Hinton. “Visualizing High-Dimensional Data Using t-SNE.” Journal of Machine Learning Research 9(Nov):2579-2605, 2008.
[2] L.J.P van der Maaten. “Accelerating t-SNE using tree-based algorithms.” Journal of Machine Learning Research 15.1:3221-3245, 2014.
[3] https://lvdmaaten.github.io/tsne/
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.