
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.

You can install the released version of campsismod from
CRAN with:
install.packages("campsismod")Alternatively, the package can also be installed with
devtools:
devtools::install_github("Calvagone/campsismod")Load 2-compartment PK model from built-in model library:
library(campsismod)
model <- model_suite$pk$`2cpt_fo`The model can be exported to files using write.
model %>% write(file="path_to_model_folder")In this case, the model code will be contained in the
model.campsis file. Parameters (THETA, OMEGA and SIGMA)
will be stored in their respective CSV file.
list.files("path_to_model_folder")
#> [1] "model.campsis" "omega.csv" "sigma.csv" "theta.csv"Alternatively, the model can also be exported in JSON format into a single file:
model %>% write(file="my_model.json")The model can be loaded from the previously created folder:
model <- read.campsis(file="path_to_model_folder")Or, from the previously created JSON file:
model <- read.campsis(file="my_model.json")The model can then be output in the console using
show:
show(model)
#> [MAIN]
#> TVBIO=THETA_BIO
#> TVKA=THETA_KA
#> TVVC=THETA_VC
#> TVVP=THETA_VP
#> TVQ=THETA_Q
#> TVCL=THETA_CL
#>
#> BIO=TVBIO
#> KA=TVKA * exp(ETA_KA)
#> VC=TVVC * exp(ETA_VC)
#> VP=TVVP * exp(ETA_VP)
#> Q=TVQ * exp(ETA_Q)
#> CL=TVCL * exp(ETA_CL)
#>
#> [ODE]
#> d/dt(A_ABS)=-KA*A_ABS
#> d/dt(A_CENTRAL)=KA*A_ABS + Q/VP*A_PERIPHERAL - Q/VC*A_CENTRAL - CL/VC*A_CENTRAL
#> d/dt(A_PERIPHERAL)=Q/VC*A_CENTRAL - Q/VP*A_PERIPHERAL
#>
#> [F]
#> A_ABS=BIO
#>
#> [ERROR]
#> CONC=A_CENTRAL/VC
#> if (CONC <= 0.001) CONC=0.001
#> CONC_ERR=CONC*(1 + EPS_PROP_RUV)
#>
#>
#> THETA's:
#> name index value fix label unit
#> 1 BIO 1 1 FALSE Bioavailability <NA>
#> 2 KA 2 1 FALSE Absorption rate 1/h
#> 3 VC 3 10 FALSE Volume of central compartment L
#> 4 VP 4 40 FALSE Volume of peripheral compartment L
#> 5 Q 5 20 FALSE Inter-compartment flow L/h
#> 6 CL 6 3 FALSE Clearance L/h
#> OMEGA's:
#> name index index2 value fix type
#> 1 KA 1 1 25 FALSE cv%
#> 2 VC 2 2 25 FALSE cv%
#> 3 VP 3 3 25 FALSE cv%
#> 4 Q 4 4 25 FALSE cv%
#> 5 CL 5 5 25 FALSE cv%
#> SIGMA's:
#> name index index2 value fix type
#> 1 PROP_RUV 1 1 0.1 FALSE sd
#> No variance-covariance matrix
#>
#> Compartments:
#> A_ABS (CMT=1)
#> A_CENTRAL (CMT=2)
#> A_PERIPHERAL (CMT=3)library(campsis)
dataset <- Dataset(5) %>%
add(Bolus(time=0, amount=1000, ii=12, addl=2)) %>%
add(Observations(times=0:36))
rxode <- model %>% simulate(dataset=dataset, dest="rxode2", seed=0)
mrgsolve <- model %>% simulate(dataset=dataset, dest="mrgsolve", seed=0)spaghettiPlot(rxode, "CONC")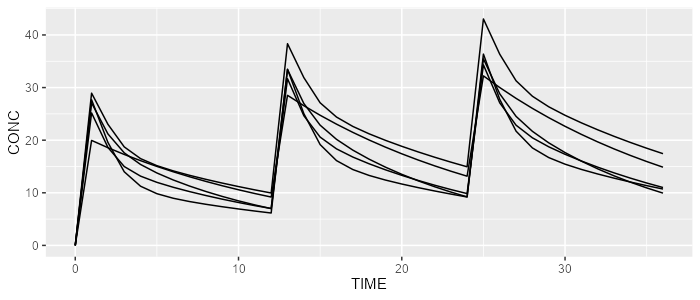
spaghettiPlot(mrgsolve, "CONC")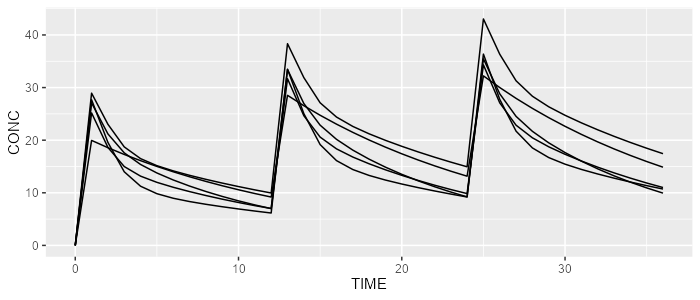
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.