
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.

charisma provides a standardized, reproducible framework for characterizing discrete color classes in digital images of biological organisms.
charisma automatically classifies colors in images into
10 human-visible categories using a biologically-inspired Color Look-Up
Table (CLUT):
Black · Blue · Brown · Green · Grey · Orange · Purple · Red · White · Yellow
✨ Fully Reproducible: Complete provenance tracking of all operations
🎨 10 Color Classes: Biologically-relevant discrete color categories
🔧 Flexible Workflows: From fully automated to completely manual
📊 Evolutionary Integration: Seamless compatibility
with geiger, phytools, pavo
⚡ High-Throughput: Designed for analyzing large image datasets
🔍 Validated CLUT: Non-overlapping HSV color space partitions
charisma depends on spatial R packages that require
system-level libraries. Install these first:
macOS (via Homebrew):
brew install udunits gdal proj geosUbuntu/Debian:
sudo apt-get install libudunits2-dev libgdal-dev libgeos-dev libproj-devFedora/RedHat:
sudo dnf install udunits2-devel gdal-devel geos-devel proj-devel# install.packages("remotes")
remotes::install_github("shawntz/charisma")install.packages("charisma") # Coming soon!library(charisma)
# Load example image
img <- system.file(
"extdata",
"Tangara_fastuosa_LACM60421.png",
package = "charisma"
)
# Basic analysis
result <- charisma(img, threshold = 0.05)
# Visualize
plot(result)
# Interactive mode with manual curation
result_interactive <- charisma(
img,
interactive = TRUE,
threshold = 0.0
)
# Save outputs
result_saved <- charisma(
img,
threshold = 0.05,
logdir = file.path(tempdir(), "charisma_outputs")
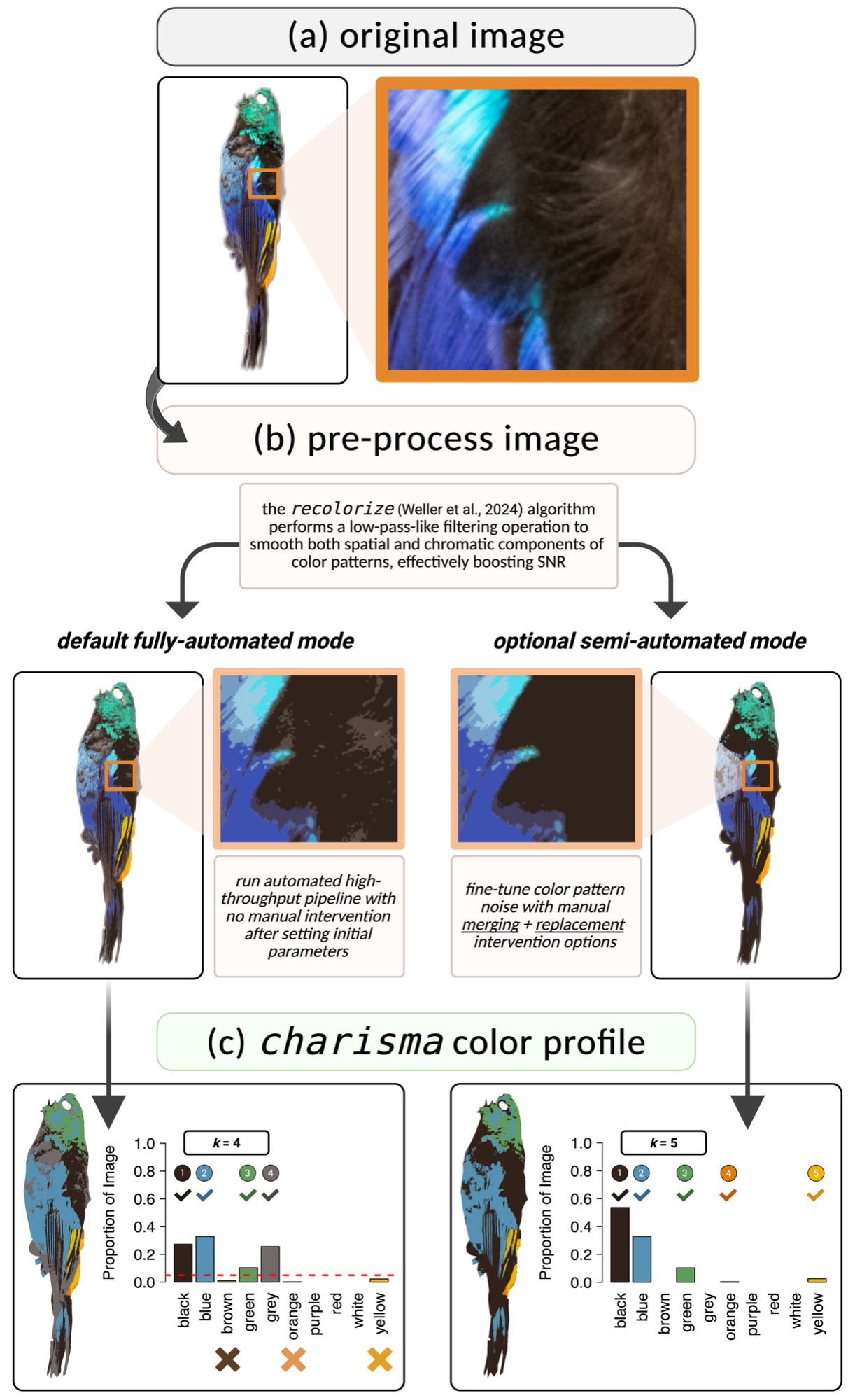
)The charisma pipeline consists of three stages:
Uses recolorize
to perform spatial-color binning, removing noisy pixels and creating a
smoothed representation of dominant colors.
Converts RGB cluster centers to HSV and matches against the CLUT
using color2label():
color2label(c(255, 0, 0)) # "red"
color2label(c(0, 0, 255)) # "blue"
color2label(c(255, 255, 0)) # "yellow"In interactive mode:
c(2,3))The charisma object contains:
# Load previous analysis
obj <- system.file("extdata", "Tangara_fastuosa.RDS", package = "charisma")
obj <- readRDS(obj)
# Apply different threshold
result2 <- charisma2(
obj,
new.threshold = 0.10
)
# Revert to specific state
result3 <- charisma2(
obj,
which.state = "merge",
state.index = 2
)# Create custom CLUT
my_clut <- charisma::clut # Start with default
# ... modify HSV ranges ...
# Validate completeness
validate(clut = my_clut)
# Use in analysis
result <- charisma(img, clut = my_clut)# Batch process images
results <- lapply(image_paths, function(img) {
charisma(img, threshold = 0.05)
})
# Extract color presence/absence
color_matrix <- do.call(rbind, lapply(results, summarize))
# Phylogenetic analyses with geiger
library(geiger)
fit_er <- fitDiscrete(
phylogeny,
color_matrix[, "blue"],
model = "ER"
)
fit_ard <- fitDiscrete(
phylogeny,
color_matrix[, "blue"],
model = "ARD"
)If you use charisma in your research, please cite:
Schwartz, S.T., Tsai, W.L.E., Karan, E.A., Juhn, M.S., Shultz, A.J., McCormack, J.E., Smith, T.B., and Alfaro, M.E. (2025). charisma: An R package to perform reproducible color characterization of digital images for biological studies. (In Review).
📧 Email: shawn.t.schwartz@gmail.com
charisma integrates with:
recolorize
(Weller et al. 2024) -
Image preprocessingpavo (Maia et al. 2019) -
Color pattern geometryMIT © 2025 Shawn T. Schwartz
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.