The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
Are you comfortable writing Surv(time, status) ~ strata
but hesitant to dive into the Fine-Gray models or custom
ggplot2 code? cifmodeling helps
clinicians, epidemiologists, and applied researchers move from basic
Kaplan-Meier curves to clear, publication-ready survival and competing
risk plots – with just a few lines of R.
It provides a unified, high-level interface for survival and competing risks analysis, combining nonparametric estimation, regression modeling, and visualization. It is centered around three tightly connected functions:
cifplot() generates a survival or cumulative incidence
function (CIF) curve. The visualization is built on top of
ggsurvfit and ggplot2.cifpanel() creates multi-panel displays for
survival/CIF curves, arranged either in a grid layout or as an
inset overlay.polyreg() fits coherent regression
models on all cause-specific CIFs simultaneously to estimate
RR/OR/SHR, offering a practical complement to Fine-Gray.Explore the main features visually:
See the Gallery for a curated set of examples usingcifplot()andcifpanel().
Learn the modeling framework with
polyreg():
See the Direct polytomous regression for coherent, joint modeling of all cause-specific CIFs.
Prefer to build intuition before diving into code?
Visit the Coffee and Research – Story and Quiz series for narrative-style introductions to study design, survival and competing risks analysis, and frequentist and causality thinking: https://gestimation.github.io/coffee-and-research/en/
library(cifmodeling)
data(diabetes.complications)
cifplot(Event(t,epsilon)~fruitq, data=diabetes.complications,
outcome.type="competing-risk", panel.per.event=TRUE)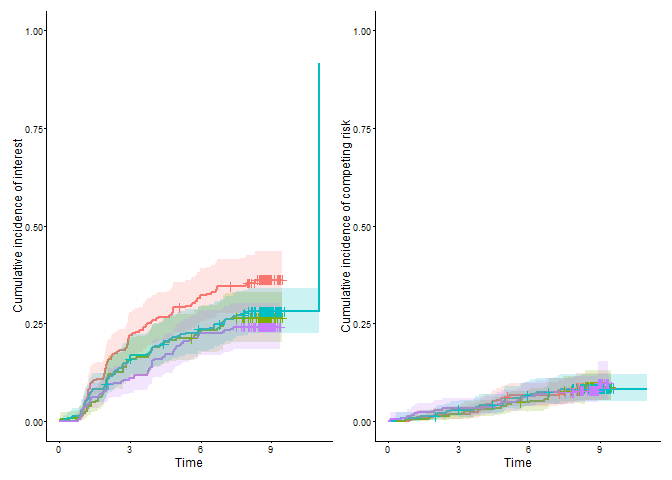
Aalen-Johansen cumulative incidence curves from cifplot()
In competing risks data, censoring is often coded as 0, the event of
interest as 1, and competing risks as 2. In the
diabetes.complications data frame, epsilon
follows this convention. With panel.per.event = TRUE,
cifplot() visualizes both competing events, with the CIF of
diabetic retinopathy (epsilon = 1) shown on the left and
the CIF of macrovascular complications (epsilon = 2) on the
right.
Four code snippets illustrate how the three core functions interact with existing resources and analyze complex competing risks data:
We use the diabetes.complications data throughout,
focusing on diabetic retinopathy (event 1) and macrovascular
complications (event 2), with the level of fruit intake, low (Q1) and
high (Q2 to 4), as the exposure (fruitq1).
The first snippet is plotting the CIF of diabetic retinopathy with
marks indicating individuals who experienced macrovascular
complications first
(add.competing.risk.mark = TRUE). Here we show a workflow
slightly different from the code at the beginning. First, the time
points at which the macrovascular complications occurred were obtained
as output1 for each strata using a helper function
extract_time_to_event(). Then, cifplot() is
used to generate the figure. The label.y,
label.x, label.strata and limit.x
arguments are also used to customize the labels and axis
limits.
data(diabetes.complications)
output1 <- extract_time_to_event(Event(t,epsilon)~fruitq1,
data=diabetes.complications, which.event="event2")
cifplot(Event(t,epsilon)~fruitq1, data=diabetes.complications,
outcome.type="competing-risk",
add.conf=FALSE, add.risktable=FALSE, add.censor.mark=FALSE,
add.competing.risk.mark=TRUE, competing.risk.time=output1,
label.y="CIF of diabetic retinopathy", label.x="Years from registration",
limits.x=c(0,8), label.strata=c("High intake","Low intake"),
level.strata=c(0, 1), order.strata=c(0, 1))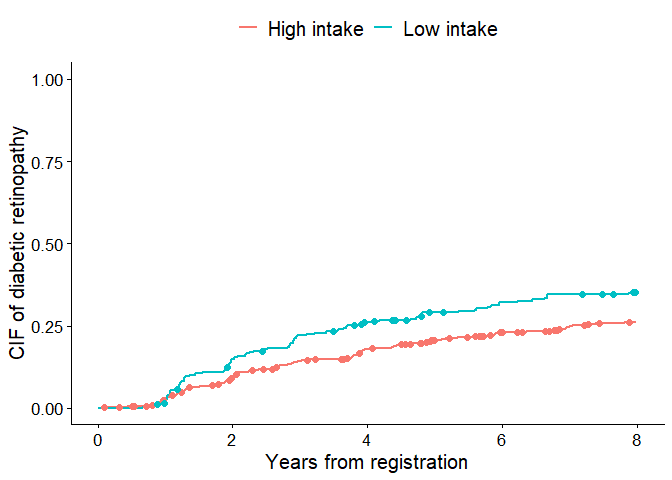
Cumulative incidence curves with competing-risk marks
In an observational study, it is necessary to adjust for confounders
in order to compare fruit intake levels. In the next example, we obtain
IPWs from CBPS() and draw adjusted CIFs. It is important to
note that in IPW analysis, the SEs output in the unweighted analysis are
not necessarily valid. Among the SE methods selectable in
cifplot() (Aalen-, delta-, and influence-function-type),
simulations by Deng and Wang (2025) have shown that the
influence-function based SE performs well. This snippet displays valid
CIs in the plot by specifying error=“if” and
add.conf=TRUE.
if (requireNamespace("CBPS", quietly = TRUE)) {
library(CBPS)
output2 <- CBPS(
fruitq1 ~ age + sex + bmi + hba1c + diabetes_duration + drug_oha + drug_insulin
+ sbp + ldl + hdl + tg + current_smoker + alcohol_drinker + ltpa,
data = diabetes.complications, ATT=0
)
diabetes.complications$ipw <- output2$weights
cifplot(Event(t,epsilon)~fruitq1, data=diabetes.complications,
outcome.type="competing-risk", weights = "ipw",
add.conf=TRUE, add.risktable=FALSE, add.censor.mark=FALSE,
label.y="CIF of diabetic retinopathy", label.x="Years from registration",
limits.x=c(0,8), label.strata=c("High intake","Low intake"),
level.strata=c(0, 1), order.strata=c(0, 1), error = "if")
} else {
plot.new()
text(0.5, 0.5,
"Install the 'CBPS' package to run this example.",
cex = 0.9)
}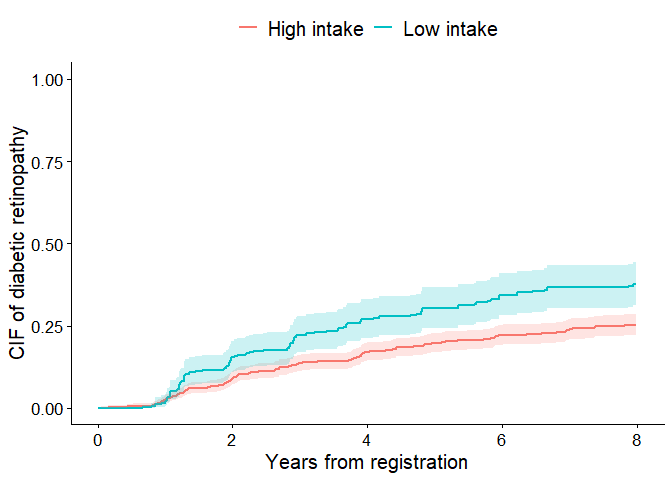
Adjusted cumulative incidence curves with CIs based on influence functions
We then fit polyreg() to estimate risk ratios for
diabetic retinopathy and macrovascular complications, in a coherent
joint model of all cause-specific CIFs at 8 years.
output3 <- polyreg(nuisance.model=Event(t,epsilon)~1, exposure="fruitq1",
data=diabetes.complications, effect.measure1="RR", effect.measure2="RR",
time.point=8, outcome.type="competing-risk")
summary(output3)
#>
#> event1 event2
#> ----------------------------------------------
#> fruitq1, 1 vs 0
#> 1.350 1.079
#> [1.114, 1.637] [0.682, 1.707]
#> (p=0.002) (p=0.746)
#>
#> ----------------------------------------------
#>
#> effect.measure RR at 8 RR at 8
#> n.events 279 in N = 978 79 in N = 978
#> median.follow.up 8 -
#> range.follow.up [0.05, 11.00] -
#> n.parameters 4 -
#> converged.by Converged in objective function -
#> nleqslv.message Function criterion near zero -The summary() method prints an event-wise table of point
estimates, CIs, and p-values. Internally, a "polyreg"
object also supports the generics API:
tidy(): coefficient-level summaries (one row per term
and per event),glance(): model-level summaries (follow-up,
convergence, number of events),augment(): observation-level diagnostics (weights for
IPCW, predicted CIFs, and influence functions).Finally, we repeat the model at multiple time points and use
modelplot() to visualize how risk ratios evolve over
follow-up. With generics-Compatible objects, polyreg() is
integrated naturally with the broader broom/modelsummary
ecosystem. For publication-ready tables, you can pass
polyreg objects directly to
modelsummary::msummary() and
modelsummary::modelplot(), including exponentiated
summaries (risk ratios, odds ratios, subdistribution hazard ratios) via
the exponentiate = TRUE option.
if (requireNamespace("modelsummary", quietly = TRUE)) {
library(modelsummary)
output4 <- polyreg(nuisance.model=Event(t,epsilon)~1, exposure="fruitq1",
data=diabetes.complications, effect.measure1="RR", effect.measure2="RR",
time.point=2, outcome.type="competing-risk")
output5 <- polyreg(nuisance.model=Event(t,epsilon)~1, exposure="fruitq1",
data=diabetes.complications, effect.measure1="RR", effect.measure2="RR",
time.point=4, outcome.type="competing-risk")
output6 <- polyreg(nuisance.model=Event(t,epsilon)~1, exposure="fruitq1",
data=diabetes.complications, effect.measure1="RR", effect.measure2="RR",
time.point=6, outcome.type="competing-risk")
summary <- list(
"RR of diabetic retinopathy at 2 years" = output4$summary$event1,
"RR of diabetic retinopathy at 4 years" = output5$summary$event1,
"RR of diabetic retinopathy at 6 years" = output6$summary$event1,
"RR of diabetic retinopathy at 8 years" = output3$summary$event1,
"RR of macrovascular complications at 2 years" = output4$summary$event2,
"RR of macrovascular complications at 4 years" = output5$summary$event2,
"RR of macrovascular complications at 6 years" = output6$summary$event2,
"RR of macrovascular complications at 8 years" = output3$summary$event2
)
modelplot(summary, coef_rename="", exponentiate = TRUE)
} else {
plot.new()
text(0.5, 0.5,
"Install the 'modelsummary' package to run this example.",
cex = 0.9)
}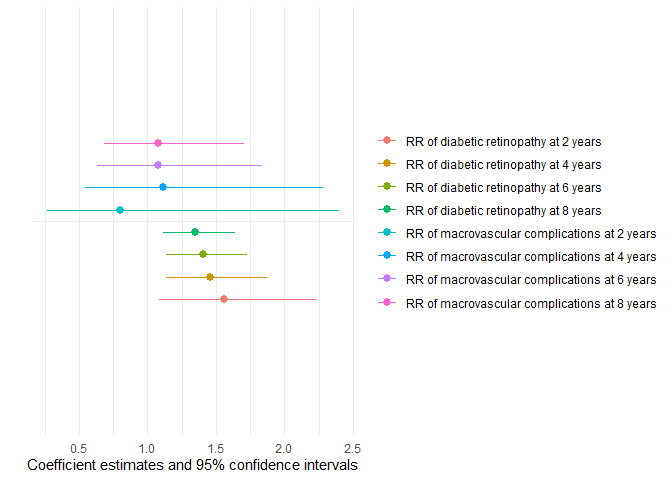
Visualizaton of risk ratios at 2, 4, 6 and 8 years using polyreg() and modelplot()
In clinical and epidemiologic research, analysts often need to handle censoring, competing risks, and intercurrent events (e.g. treatment switching), but existing R packages typically separate these tasks across different interfaces. cifmodeling provides a unified, publication-ready toolkit that integrates nonparametric estimation, regression modeling, and visualization for survival and competing risks data. The tools assist users in the following ways:
Event() + formula + data syntax.polyreg() directly targets
ratios of CIFs (risk ratios, odds ratios, subdistribution hazard
ratios), so parameters align closely with differences seen in CIF
curves.polyreg() models all cause-specific CIFs together,
parameterizing the nuisance structure with polytomous log odds products
and enforcing that their CIFs sum to at most one.generics::tidy(), glance(), and
augment(), which integrate polyreg() smoothly
with modelsummary and other broom-style tools.ggsurvfit and ggplot2, including
number-at-risk/CIF+CI tables,
censoring/competing-risk/intercurrent-event marks, and multi-panel
layouts.Several excellent R packages exist for survival and competing risks
analysis. The survival package provides the canonical
API for survival data. In combination with the
ggsurvfit package, survival::survfit() can
produce publication-ready survival plots. For CIF plots, however,
integration in the general ecosystem is less streamlined.
cifmodeling fills this gap by offering
cifplot() for survival/CIF plots and multi-panel figures
via a single, unified interface.
Beyond providing a unified interface, cifcurve() also
extends survfit() in a few targeted ways. For unweighted
survival data, it reproduces the standard Kaplan-Meier estimator with
Greenwood and Tsiatis SEs and a unified set of CI
transformations. For competing risks data, it computes Aalen-Johansen
CIFs with both Aalen-type and delta-method SEs. For
weighted survival or competing risks data (e.g. inverse probability
weighting), it implements influence-function based SEs
(Deng and Wang 2025) as well as modified Greenwood- and
Tsiatis-type SEs (Xie and Liu 2005), which are valid under
general positive weights.
If you need very fine-grained plot customization, you can compute the
estimator and keep a survfit-compatible object with
cifcurve() (or supply your own survfit object) and then
style it using ggsurvfit/ggplot2 layers. In other
words:
cifcurve() for estimation,cifplot() / cifpanel() for quick,
high-quality figures, andggplot ecosystem when you want full
artistic control.The causalRisk package offers IPW-based estimation of counterfactual cumulative risks and hazards. It is most relevant when treatment, censoring, and missingness mechanisms must be modeled explicitly. In contrast, cifmodeling focuses on nonparametric estimation and direct CIF regression rather than structural causal models.
The mets package is a more specialized toolkit that
provides advanced methods for competing risks analysis.
cifmodeling::polyreg() focuses on coherent modeling of all
CIFs simultaneously to estimate the exposure effects in terms of
RR/OR/SHR. This coherence can come with longer runtimes for large
problems. If you prefer fitting separate regression models for each
competing event or specifically need the Fine-Gray models (Fine and Gray
1999) and the direct binomial models (Scheike, Zhang and Gerds 2008),
mets::cifreg() and mets::binreg() are
excellent choices.
In short, cifmodeling provides a unified high-level grammar for estimation, visualization, and direct CIF regression — something no existing package currently offers in one place.
Interested in the precise variance formulas and influence functions for the Aalen-Johansen estimator?
Visit Computational formulas in cifcurve().
| Function | cifmodeling | survival | cmprsk | mets | ggsurvfit |
|---|---|---|---|---|---|
| AJ estimator | Yes | Yes (multistate survfit) |
Yes (cuminc) |
Yes | Depends on input |
| Weighted AJ estimator and valid SE | Yes (IPW + IF-based SE) | Yes (case weights / robust SE) | No | Yes (IPW + IF-based SE) | Depends on input |
| Gray test | No | No (only log-rank via survdiff) |
Yes (cuminc) |
No | tidycmprsk::glance() + add_pvalue() |
| Fine–Gray model | No | finegray+coxph |
crr |
cifregFG |
No |
| Direct CIF regression | polyreg |
No | No | cifreg, binreg |
No |
| Surv()/Event() interface | Yes (Event, Surv) |
Yes (Surv) |
No (ftime/fstatus) |
Yes (Event, Surv) |
Yes (Surv, ggcuminc + tidiers) |
| Publication-ready survival/CIF plot | Yes (cifplot, cifpanel) |
plot (base) |
plot.cuminc (base) |
plot (base) |
Designed for publication |
| Support for tidy/glance/augment | Yes (polyreg + methods) |
Yes (via broom) | Yes (via tidycmprsk) | No | Yes (via broom) |
The package is implemented in R and relies on Rcpp,
nleqslv and boot for its numerical back-end.
The examples in this document also use ggplot2,
ggsurvfit, patchwork and
modelsummary for tabulation and plotting. Install the core
package and these companion packages with:
# Install cifmodeling with core dependencies
install.packages(c("cifmodeling", "Rcpp", "nleqslv", "boot"))
# Recommended packages for plotting and tabulation in this README
install.packages(c("ggplot2", "ggsurvfit", "patchwork", "modelsummary"))cifmodeling includes an extensive test suite built
with testthat, which checks the numerical accuracy and
graphical consistency of all core functions (cifcurve(),
cifplot(), cifpanel(), and
polyreg()). The estimators are routinely compared against
related functions in survival, cmprsk
and mets packages to ensure consistency. The package is
continuously tested on GitHub Actions (Windows, macOS, Linux) to
maintain reproducibility and CRAN-level compliance.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.