
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.


The clinicalsignificance R
package provides a comprehensive toolkit for analyzing clinical
significance in intervention studies.
Why this package? While statistical significance asks: “Is this effect unlikely due to chance?” Clinical significance asks: “Does this intervention make a meaningful difference for the patient?”
This package empowers researchers and practitioners to move beyond p-values and assess the practical relevance of treatment outcomes.
The package implements the most widely used methods for clinical significance analysis. Each approach answers a specific question:
cs_anchor(): Did the patient
improve by a minimal amount? Evaluates change based on a predefined
Minimal Important Difference (MID).cs_percentage(): Did the patient
improve by a certain percentage? Assesses change relative to the
baseline score.cs_distribution(): Is the change
reliable (beyond measurement error)? Uses statistical distribution
metrics like the Reliable Change Index (RCI).cs_statistical(): Did the patient
return to a functional range? Determines if a patient moved from a
clinical to a functional population.cs_combined(): The “Gold Standard”
(e.g., Jacobson & Truax). Combines reliability and cutoff
criteria for a robust assessment.Install the stable version from CRAN:
install.packages("clinicalsignificance")Or the development version from GitHub:
# install.packages("pak")
pak::pak("benediktclaus/clinicalsignificance")Let’s look at the combined approach (Jacobson &
Truax, 1991). We want to know if patients in the claus_2020
dataset (included in the package) showed a reliable
change AND moved into a functional population
range.
library(clinicalsignificance)
library(ggplot2)
# 1. Perform the analysis
results_combined <- claus_2020 |>
cs_combined(
id = id,
time = time,
outcome = bdi,
pre = 1,
post = 4,
reliability = 0.801,
m_functional = 7.69,
sd_functional = 7.52,
cutoff_type = "c"
)
# 2. Visualize the results
plot(results_combined, show_group = "category")
#> Ignoring unknown labels:
#> • colour : "Group"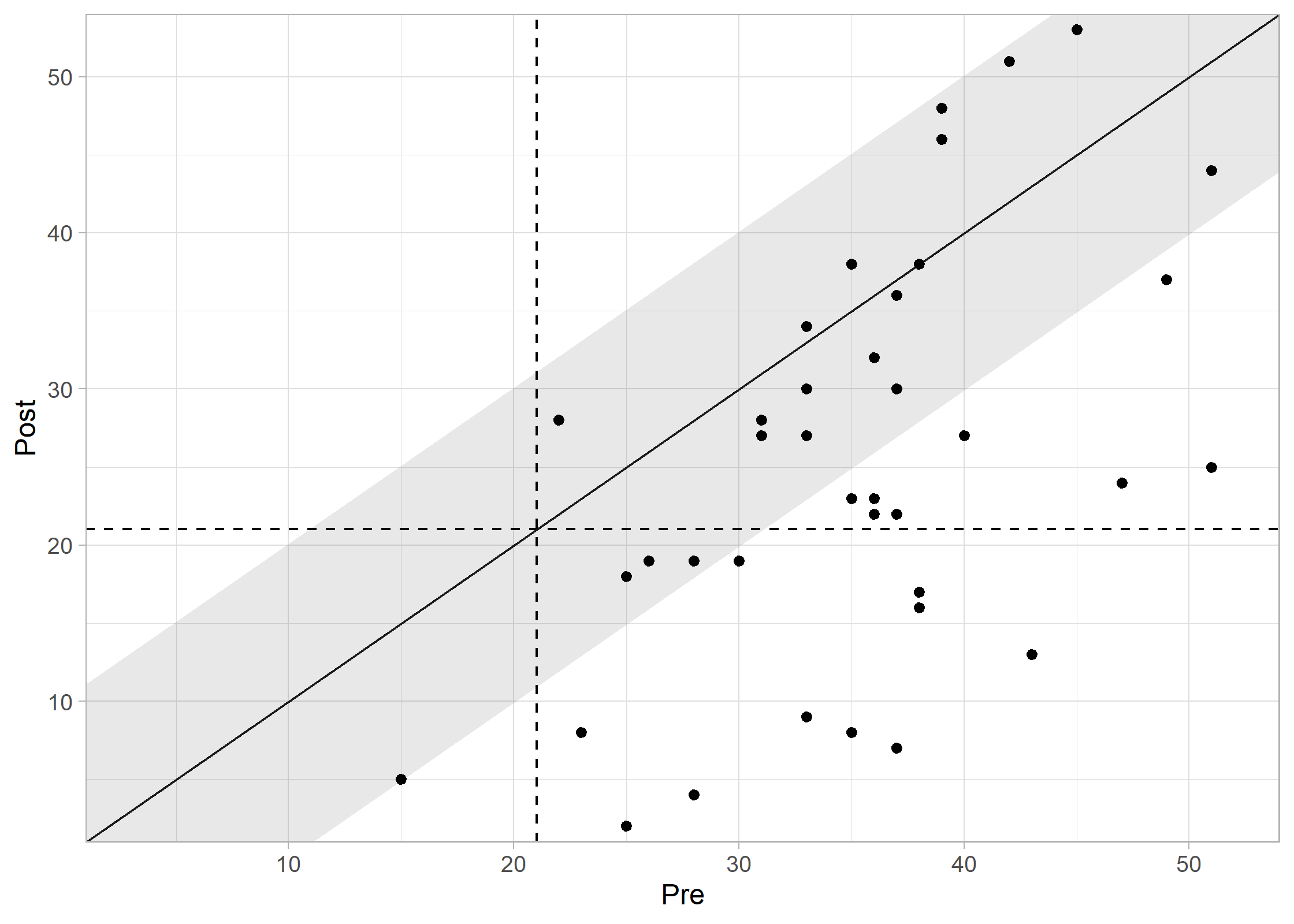
Interpreting the Plot: * Recovered (Green): Reliable improvement + moved to functional range. * Improved (Blue): Reliable improvement, but still in clinical range. * Unchanged (Grey): No reliable change. * Deteriorated (Red): Reliable worsening.
# 3. Get a summary table
summary(results_combined)
#>
#> ---- Clinical Significance Results ----
#>
#> Approach: Distribution-based
#> RCI Method: JT
#> N (original): 43
#> N (used): 40
#> Percent used: 93.02%
#> Outcome: bdi
#> Cutoff Type: c
#> Cutoff: 21.02
#> Outcome: bdi
#> Reliability: 0.801
#>
#> -- Cutoff Descriptives
#>
#> M Clinical | SD Clinical | M Functional | SD Functional
#> -------------------------------------------------------
#> 35.48 | 8.16 | 7.69 | 7.52
#>
#>
#> -- Results
#>
#> Category | N | Percent
#> ---------------------------
#> Recovered | 10 | 25.00%
#> Improved | 8 | 20.00%
#> Unchanged | 22 | 55.00%
#> Deteriorated | 0 | 0.00%
#> Harmed | 0 | 0.00%Please cite both the package and the JSS paper if you use
clinicalsignificance in your research.
Claus, B. B., Wager, J., & Bonnet, U. (2024). clinicalsignificance: Clinical Significance Analyses of Intervention Studies in R. Journal of Statistical Software, 111(1), 1–39. https://doi.org/10.18637/jss.v111.i01
@article{JSS:v111:i01,
author = {Benedikt B. Claus and Julia Wager and Udo Bonnet},
title = {{clinicalsignificance}: Clinical Significance Analyses of Intervention Studies in {R}},
journal = {Journal of Statistical Software},
year = {2024},
volume = {111},
number = {1},
pages = {1--39},
doi = {10.18637/jss.v111.i01},
}
@manual{R-clinicalsignificance,
title = {clinicalsignificance: A Toolbox for Clinical Significance Analyses in Intervention Studies},
author = {Benedikt B. Claus},
year = {2024},
note = {R package version 2.1.0},
doi = {10.32614/CRAN.package.clinicalsignificance},
url = {[https://github.com/benediktclaus/clinicalsignificance/](https://github.com/benediktclaus/clinicalsignificance/)},
}Contributions are welcome! If you encounter bugs or have feature requests: 1. Check the Issue Tracker. 2. Submit a Pull Request.
License: GNU General Public License v3.0
Built with ❤️ for better clinical research.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.