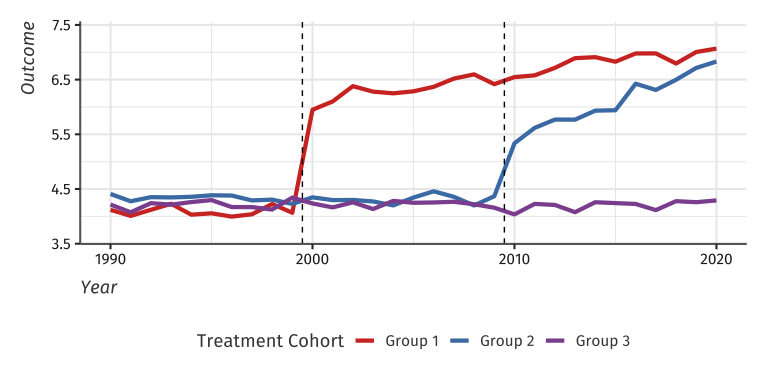
Example data with heterogeneous treatment effects
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
The goal of didimputation is to estimate TWFE models without running into the problem of staggered treatment adoption.
You can install didimputation from github with:
devtools::install_github("kylebutts/didimputation")I will load example data from the package and plot the average outcome among the groups. Here is one unit’s data:
library(didimputation)
#> Loading required package: fixest
#> Loading required package: data.table
library(fixest)
library(ggplot2)
# Load Data from did2s package
data("df_het", package = "didimputation")
setDT(df_het)Here is a plot of the average outcome variable for each of the groups:
# Plot Data
df_avg <- df_het[,
.(dep_var = mean(dep_var)),
by = .(group, year)
]
# Get treatment years for plotting
gs <- df_het[treat == TRUE, unique(g)]
ggplot() +
geom_line(data = df_avg, mapping = aes(y = dep_var, x = year, color = group), size = 1.5) +
geom_vline(xintercept = gs - 0.5, linetype = "dashed") +
theme_minimal(base_size = 16) +
theme(legend.position = "bottom") +
labs(y = "Outcome", x = "Year", color = "Treatment Cohort") +
scale_y_continuous(expand = expansion(add = .5)) +
scale_color_manual(values = c("Group 1" = "#d2382c", "Group 2" = "#497eb3", "Group 3" = "#8e549f"))
#> Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` instead.
#> This warning is displayed once every 8 hours.
#> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
#> generated.
Example data with heterogeneous treatment effects
First, lets estimate a static did:
# Static
static <- did_imputation(data = df_het, yname = "dep_var", gname = "g", tname = "year", idname = "unit")
static
#> term estimate std.error conf.low conf.high
#> <char> <num> <num> <num> <num>
#> 1: treat 2.262952 0.03139684 2.201414 2.32449This is very close to the true treatment effect of 2.2384912.
Then, let’s estimate an event study did:
# Event Study
es <- did_imputation(
data = df_het, yname = "dep_var", gname = "g",
tname = "year", idname = "unit",
# event-study
horizon = TRUE, pretrends = -5:-1
)
es
#> term estimate std.error conf.low conf.high
#> <char> <num> <num> <num> <num>
#> 1: -5 -0.06412085 0.07634962 -0.21376611 0.08552441
#> 2: -4 -0.01201577 0.07634962 -0.16166103 0.13762949
#> 3: -3 -0.01387197 0.07634962 -0.16351723 0.13577329
#> 4: -2 0.05103140 0.07634962 -0.09861386 0.20067666
#> 5: -1 0.02022464 0.07634962 -0.12942062 0.16986990
#> 6: 0 1.51314201 0.07547736 1.36520639 1.66107763
#> 7: 1 1.66384318 0.07675141 1.51341041 1.81427594
#> 8: 2 1.86436720 0.07450151 1.71834424 2.01039015
#> 9: 3 1.91872093 0.07471704 1.77227552 2.06516633
#> 10: 4 1.87322387 0.07418170 1.72782773 2.01862001
#> 11: 5 1.87844597 0.07567190 1.73012905 2.02676290
#> 12: 6 2.14373139 0.07632691 1.99413065 2.29333213
#> 13: 7 2.23777696 0.07610842 2.08860445 2.38694946
#> 14: 8 2.33650066 0.07446268 2.19055381 2.48244751
#> 15: 9 2.34352836 0.07471679 2.19708345 2.48997326
#> 16: 10 2.53443351 0.08109550 2.37548633 2.69338068
#> 17: 11 2.47944533 0.11953547 2.24515580 2.71373486
#> 18: 12 2.63493727 0.11531779 2.40891439 2.86096014
#> 19: 13 2.94449757 0.11047299 2.72797052 3.16102462
#> 20: 14 2.78171206 0.11466367 2.55697127 3.00645285
#> 21: 15 2.71470743 0.12030494 2.47890975 2.95050510
#> 22: 16 2.88065382 0.11563154 2.65401601 3.10729163
#> 23: 17 2.99383855 0.11438496 2.76964404 3.21803306
#> 24: 18 2.64616896 0.11545789 2.41987148 2.87246643
#> 25: 19 2.87530636 0.11405840 2.65175189 3.09886082
#> 26: 20 2.90465651 0.11320219 2.68278023 3.12653280
#> term estimate std.error conf.low conf.highAnd plot the results:
pts <- es |>
as.data.table() |>
DT(, .(rel_year = term, estimate, std.error)) |>
DT(, let(
ci_lower = estimate - 1.96 * std.error,
ci_upper = estimate + 1.96 * std.error,
group = "DID Imputation Estimate",
rel_year = as.numeric(rel_year)
))
te_true <- df_het |>
DT(
g > 0,
.(estimate = mean(te + te_dynamic)),
by = "rel_year"
) |>
DT(, group := "True Effect")
pts <- rbind(pts, te_true, fill = TRUE)
pts <- pts |>
DT(rel_year >= -5 & rel_year <= 7, ) |>
DT(, rel_year := ifelse(group == "DID Imputation Estimate", rel_year - 0.1, rel_year))
max_y <- max(pts$estimate)
ggplot() +
# 0 effect
geom_hline(yintercept = 0, linetype = "dashed") +
geom_vline(xintercept = -0.5, linetype = "dashed") +
# Confidence Intervals
geom_linerange(data = pts, mapping = aes(x = rel_year, ymin = ci_lower, ymax = ci_upper), color = "grey30") +
# Estimates
geom_point(data = pts, mapping = aes(x = rel_year, y = estimate, color = group), size = 2) +
# Label
geom_label(
data = data.frame(x = -0.5 - 0.1, y = max_y + 0.25, label = "Treatment Starts ▶"), label.size = NA,
mapping = aes(x = x, y = y, label = label), size = 5.5, hjust = 1, fontface = 2, inherit.aes = FALSE
) +
scale_x_continuous(breaks = -8:8, minor_breaks = NULL) +
scale_y_continuous(minor_breaks = NULL) +
scale_color_manual(values = c("DID Imputation Estimate" = "steelblue", "True Effect" = "#b44682")) +
labs(x = "Relative Time", y = "Estimate", color = NULL, title = NULL) +
theme_minimal(base_size = 16) +
theme(legend.position = "bottom")
#> Warning: Removed 13 rows containing missing values (`geom_segment()`).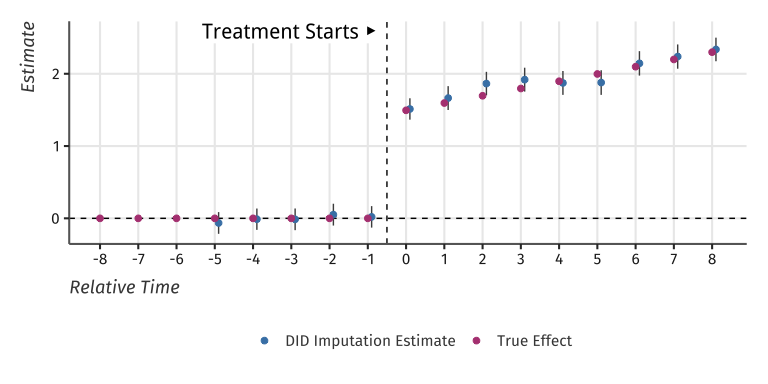
Event-study plot with example data
# TWFE
twfe <- fixest::feols(dep_var ~ i(rel_year, ref = c(-1, Inf)) | unit + year, data = df_het)
twfe_est <- broom::tidy(twfe)
twfe_est <- twfe_est |>
DT(grepl("rel_year::", term)) |>
DT(, .(rel_year = term, estimate, std.error)) |>
DT(, let(
rel_year = as.numeric(gsub("rel_year::", "", rel_year)),
ci_lower = estimate - 1.96 * std.error,
ci_upper = estimate + 1.96 * std.error,
group = "TWFE Estimate"
)) |>
DT(rel_year >= -5 & rel_year <= 7, ) |>
DT(, rel_year := rel_year + 0.1)
# Add TWFE Points
both_pts <- rbind(pts, twfe_est, fill = TRUE)
max_y <- max(pts$estimate)
ggplot() +
# 0 effect
geom_hline(yintercept = 0, linetype = "dashed") +
geom_vline(xintercept = -0.5, linetype = "dashed") +
# Confidence Intervals
geom_linerange(data = both_pts, mapping = aes(x = rel_year, ymin = ci_lower, ymax = ci_upper), color = "grey30") +
# Estimates
geom_point(data = both_pts, mapping = aes(x = rel_year, y = estimate, color = group), size = 2) +
# Label
geom_label(
data = data.frame(x = -0.5 - 0.1, y = max_y + 0.25, label = "Treatment Starts ▶"), label.size = NA,
mapping = aes(x = x, y = y, label = label), size = 5.5, hjust = 1, fontface = 2, inherit.aes = FALSE
) +
scale_x_continuous(breaks = -8:8, minor_breaks = NULL) +
scale_y_continuous(minor_breaks = NULL) +
scale_color_manual(values = c("DID Imputation Estimate" = "steelblue", "True Effect" = "#b44682", "TWFE Estimate" = "#82b446")) +
labs(x = "Relative Time", y = "Estimate", color = NULL, title = NULL) +
theme_minimal(base_size = 16) +
theme(legend.position = "bottom")
#> Warning: Removed 13 rows containing missing values (`geom_segment()`).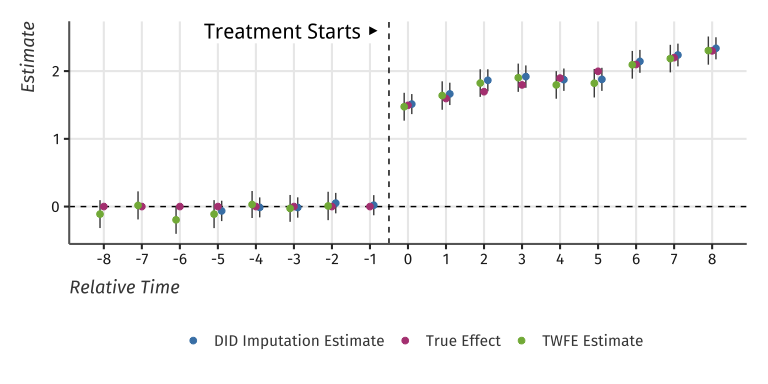
TWFE and Two-Stage estimates of Event-Study
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.