The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
The name diegr comes from Dynamic and Interactive EEG
Graphics using R. The diegr package enables
researchers to visualize high-density electroencephalography (HD-EEG)
data with animated and interactive graphics, supporting both exploratory
and confirmatory analyses of sensor-level brain signals.
The package diegr includes:
boxplot_epoch,
boxplot_subject, boxplot_rt)interactive_waveforms)topo_plot)scalp_plot)summary_stats_rt,baseline_correction,
compute_mean)pick_data, pick_region)plot_time_mean, plot_topo_mean)animate_topo, animate_topo_mean,
animate_scalp)You can install the current version of diegr from CRAN
with:
install.packages("diegr")or the latest development version from GitHub with:
# install.packages("devtools")
devtools::install_github("gerslovaz/diegr") Because of large volumes of data obtained from HD-EEG measurements, the package allows users to work directly with database tables (in addition to common formats such as data frames or tibbles). Such a procedure is more efficient in terms of memory usage.
The database you want to use as input to diegr functions
must contain columns with the following structure:
group - ID of groups,subject - ID of subjects,sensor - sensor labels,epoch - epoch numbers,condition - labels of experimental condition,time - numbers of time points (as sampling points, not
in ms),signal - the EEG signal amplitude in microvolts (in
most functions it is possible to set the name of the column containing
the amplitude arbitrarily).Note: It is not necessary for the data to contain all variables, but if it does, they must be named according to the structure presented above.
The package contains some included training datasets:
epochdata: epoched HD-EEG data (anonymized short slice
from big HD-EEG study presented in Madetko-Alster, 2025) for 2 subjects
and 204 selected sensors in 50 time points,HCGSN256: a list with Cartesian coordinates of HD-EEG
sensor positions in 3D space on the scalp surface and their projection
into 2D spacertdata: response times (time between stimulus
presentation and pressing the button) from the experiment involving a
simple visual motor task (anonymized short slice from big HD-EEG study
presented in Madetko-Alster, 2025).For more information about the structure of built-in data see the
package vignette vignette("diegr", package = "diegr").
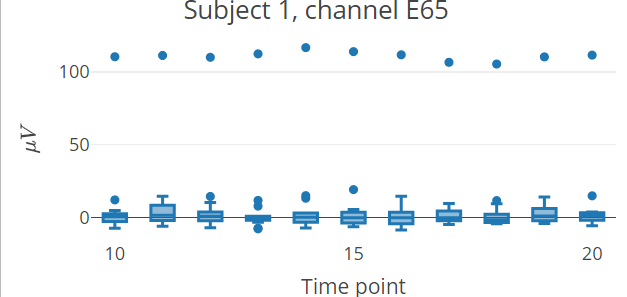
This is a basic example which shows how to plot interactive epoch boxplots from chosen electrode in different time points for one subject:
library(diegr)
data("epochdata")epochdata |>
pick_data(subject_rg = 1, sensor_rg = "E65") |>
boxplot_epoch(amplitude = "signal", time_lim = c(10:20))
Note: The README format does not allow the inclusion of
plotly interactive elements, only the static preview of the
result is shown.
data("HCGSN256")
# creating a mesh
M1 <- point_mesh(dimension = 2, n = 30000, type = "polygon", sensor_select = unique(epochdata$sensor))
# filtering a subset of data to display
data_short <- epochdata |>
pick_data(subject_rg = 1, time_rg = 15, epoch_rg = 10)
# or you can use dplyr::filter()
# dplyr::filter(subject == 1 & epoch == 10 & time == 15)
# function for displaying a topographic map of the chosen signal on the created mesh M1
topo_plot(data_short, amplitude = "signal", mesh = M1)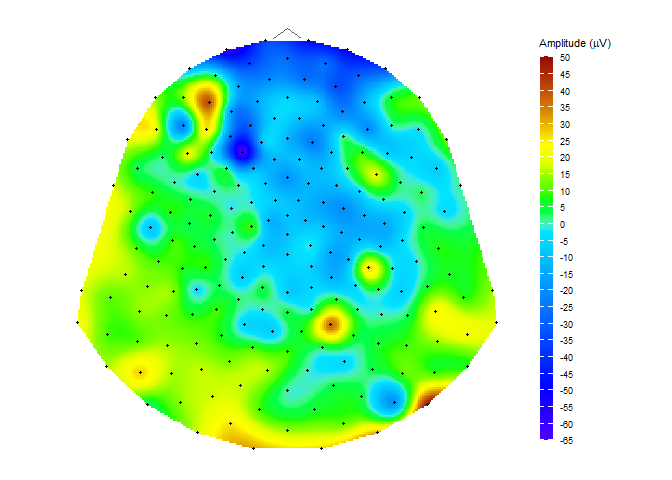
Compute the average signal for subject 2 from the channels E65 and E34 (exclude the oulier epochs 14 and 15) and then display it along with CI bounds (use plot_time_mean conditioned by sensor)
# extract required data
edata <- epochdata |>
pick_data(subject_rg = 2, sensor_rg = c("E34", "E65"), epoch_rg = 1:13)
# baseline correction
data_base <- baseline_correction(edata, baseline_range = 1:10)
# compute average
data_mean <- data_base |>
compute_mean(amplitude = "signal_base", type = "point", domain = "time")
# plot the average line with CI
plot_time_mean(data = data_mean, t0 = 10, condition_column = "sensor", legend_title = "Sensor")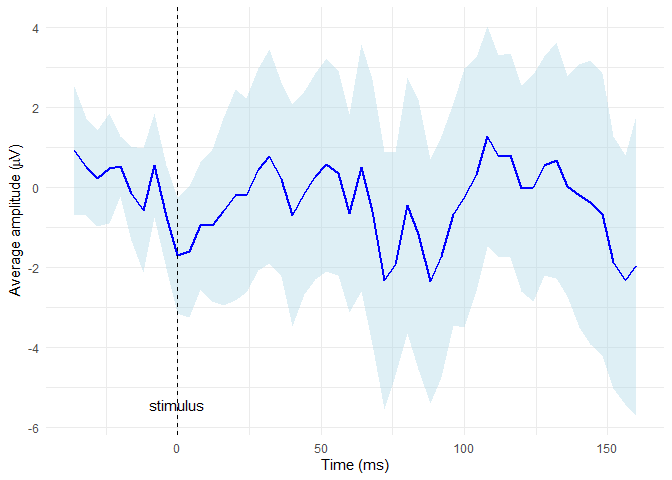
For detailed examples and usage explanation, please see the package
vignette: vignette("diegr", package = "diegr").
References Madetko-Alster N., Alster P., Lamoš M., Šmahovská L., Boušek T., Rektor I. and Bočková M. The role of the somatosensory cortex in self-paced movement impairment in Parkinson’s disease. Clinical Neurophysiology. 2025, vol. 171, 11-17. https://doi.org/10.1016/j.clinph.2025.01.001
License This package is distributed under the MIT license. See LICENSE file for details.
Citation Use citation("diegr") to cite
this package.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.