The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
The goal of eudract is to provide tools to easily produce summaries of safety data from clinical trials that can easily be uploaded into eudraCT or ClinTrials.gov .
An additional feature produces commonly used tables and figures that feature in statistical reports and papers for clinical trials.
You can install from CRAN directly with
install.packages("eudract")You can install the very latest version on github with:
install.packages("devtools")
devtools::install_github("shug0131/eudract_pkg")https://shug0131.github.io/eudraCT/ provides full documentation
Go and read the help pages within R
?eudract::safety_summary
safety_statistics <- safety_summary(safety,
exposed=c("Experimental"=60,"Control"=67))
simple <- tempfile(fileext = ".xml")
eudract <- tempfile(fileext = ".xml")
ct <- tempfile(fileext = ".xml")
simple_safety_xml(safety_statistics, simple)
eudract_convert(input=simple,
output=eudract)
clintrials_gov_convert(input=simple,
original=system.file("extdata", "1234.xml", package ="eudract"),
output=ct)
\dontrun{
# This needs a real user account to work
clintrials_gov_upload(
input=simple,
orgname="CTU",
username="Student",
password="Guinness",
studyid="1234"
)
}And for produce standard reporting outputs
library(eudract)
safety_statistics <- safety_summary(safety,
exposed=c("Control"=99, "Experimental"=101))
safety_statistics$GROUP
#> title subjectsAffectedBySeriousAdverseEvents
#> 1 Control 15
#> 2 Experimental 33
#> subjectsAffectedByNonSeriousAdverseEvents deathsResultingFromAdverseEvents
#> 1 15 9
#> 2 24 22
#> subjectsExposed deathsAllCauses
#> 1 99 9
#> 2 101 22
head( incidence_table(safety_statistics, type="serious") )
#> System Organ Class Preferred Term
#> 1 Blood and lymphatic system disorders B-cell lymphoma
#> 2 Cardiac disorders Cardiac arrest
#> 3 Cardiac failure congestive
#> 4 Gastrointestinal disorders Abdominal pain
#> 5 Gastroenteritis viral
#> 6 Gastrointestinal haemorrhage
#> Control (N = 99) Experimental (N = 101)
#> 1 1% (1, 1) 0% (0, 0)
#> 2 1% (1, 1) 0% (0, 0)
#> 3 0% (0, 0) 1% (1, 1)
#> 4 1% (1, 1) 0% (0, 0)
#> 5 0% (0, 0) 1% (1, 1)
#> 6 0% (0, 0) 1% (1, 1)
relative_risk_table(safety_statistics, type="serious")
#> System Organ Class Preferred Term Relative Risk (C.I.)
#> 1 Hepatobiliary disorders Cholecystitis acute 0.98 (0.0605, 15.9)
#> 2 Immune system disorders Vasculitis 1.96 (0.175, 22)
#> 3 Surgical and medical procedures Hip arthroplasty 1.96 (0.175, 22)
#> 4 Vascular disorders Deep vein thrombosis 2.94 (0.301, 28.8)
#> 5 Pulmonary embolism 0.98 (0.0605, 15.9)
dot_plot(safety_statistics, type="serious", base=4) 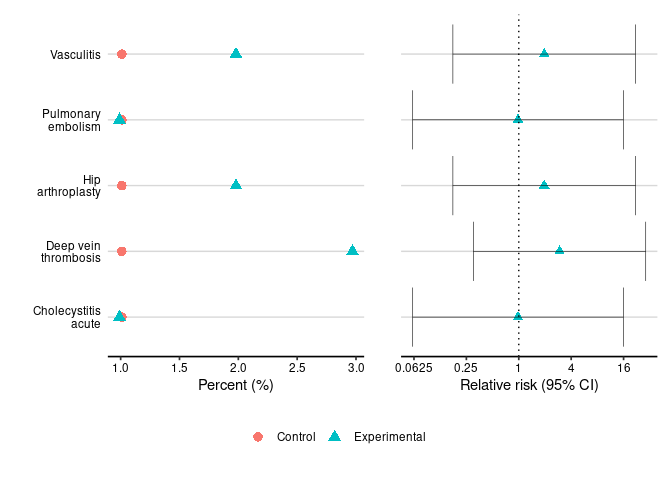
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.