
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.

We developed fullRankMatrix primarily for one-hot
encoded design matrices used in linear models. In our case, we were
faced with a 1-hot encoded design matrix, that had a lot of linearly
dependent columns. This happened when modeling a lot of interaction
terms. Since fitting a linear model on a design matrix with linearly
dependent columns will produce results that can lead to misleading
interpretation (s. example below), we decided to develop a package that
will help with identifying linearly dependent columns and replacing them
with columns constructed of orthogonal vectors that span the space of
the previously linearly dependent columns.
The goal of fullRankMatrix is to remove empty columns
(contain only 0s), merge duplicated columns (containing the same
entries) and merge linearly dependent columns. These operations will
create a matrix of full rank. The changes made to the columns are
reflected in the column headers such that the columns can still be
interpreted if the matrix is used in e.g. a linear model fit.
You can install fullRankMatrix directly from CRAN. Just
paste the following snippet into your R console:
install.packages("fullRankMatrix")You can install the development version of
fullRankMatrix from GitHub
with:
# install.packages("devtools")
devtools::install_github("Pweidemueller/fullRankMatrix")If you want to cite this package in a publication, you can run the following command in your R console:
citation("fullRankMatrix")
#> To cite package 'fullRankMatrix' in publications use:
#>
#> Weidemüller P, Ahlmann-Eltze C (2024). _fullRankMatrix: Produce a
#> Full Rank Matrix_. R package version 0.1.0,
#> <https://github.com/Pweidemueller/fullRankMatrix>.
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Manual{,
#> title = {fullRankMatrix: Produce a Full Rank Matrix},
#> author = {Paula Weidemüller and Constantin Ahlmann-Eltze},
#> year = {2024},
#> note = {R package version 0.1.0},
#> url = {https://github.com/Pweidemueller/fullRankMatrix},
#> }In order to visualize it, let’s look at a very simple example. Say we
have a matrix with three columns, each with three entries. These columns
can be visualized as vectors in a coordinate system with 3 axes (s.
image). The first vector points into the plane spanned by the first and
third axis. The second and third vectors lie in the plane spanned by the
first and second axis. Since this is a very simple example, we
immediately spot that the third column is a multiple of the second
column. Their corresponding vectors lie perfectly on top of each other.
This means instead of the two columns spanning a 2D space they just
occupy a line, i.e. a 1D space. This is identified by
fullRankMatrix, which replaces these two linearly dependent
columns with one vector that describes the 1D space in which column 2
and column 3 used to lie. The resulting matrix is now full rank with no
linearly dependent columns.
library(fullRankMatrix)c1 <- c(1, 0, 1)
c2 <- c(1, 2, 0)
c3 <- c(2, 4, 0)
mat <- cbind(c1, c2, c3)
make_full_rank_matrix(mat)
#> $matrix
#> c1 SPACE_1_AXIS1
#> [1,] 1 -0.4472136
#> [2,] 0 -0.8944272
#> [3,] 1 0.0000000
#>
#> $space_list
#> $space_list$SPACE_1
#> [1] "c2" "c3"knitr::include_graphics("man/figures/example_vectors.png")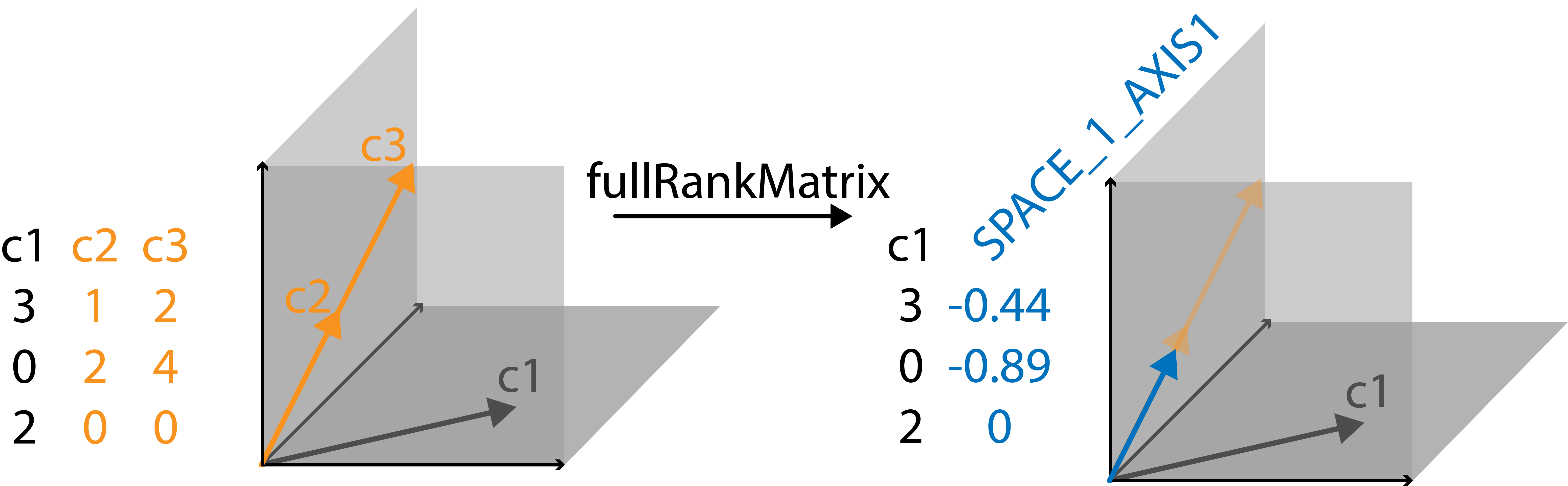
Visualisation of identifying and replacing linearly dependent columns.
Above was a rather abstract example that was easy to visualize, let’s
now walk through the utilities of fullRankMatrix when
applied to a more realistic design matrix.
When using linear models you should check if any of the columns in your design matrix are linearly dependent. If there are, this will alter the interpretation of the fit. Here is a rather constructed example where we are interested in identifying which ingredients contribute mostly to the sweetness of fruit salads.
# let's say we have 10 fruit salads and indicate which ingredients are present in each salad
strawberry <- c(1,1,1,1,0,0,0,0,0,0)
poppyseed <- c(0,0,0,0,1,1,1,0,0,0)
orange <- c(1,1,1,1,1,1,1,0,0,0)
pear <- c(0,0,0,1,0,0,0,1,1,1)
mint <- c(1,1,0,0,0,0,0,0,0,0)
apple <- c(0,0,0,0,0,0,1,1,1,1)
# let's pretend we know how each fruit influences the sweetness of a fruit salad
# in this case we say that strawberries and oranges have the biggest influence on sweetness
set.seed(30)
strawberry_sweet <- strawberry * rnorm(10, 4)
poppyseed_sweet <- poppyseed * rnorm(10, 0.1)
orange_sweet <- orange * rnorm(10, 5)
pear_sweet <- pear * rnorm(10, 0.5)
mint_sweet <- mint * rnorm(10, 1)
apple_sweet <- apple * rnorm(10, 2)
sweetness <- strawberry_sweet + poppyseed_sweet+ orange_sweet + pear_sweet +
mint_sweet + apple_sweet
mat <- cbind(strawberry,poppyseed,orange,pear,mint,apple)
fit <- lm(sweetness ~ mat + 0)
print(summary(fit))
#>
#> Call:
#> lm(formula = sweetness ~ mat + 0)
#>
#> Residuals:
#> 1 2 3 4 5 6 7 8
#> -2.00934 2.00934 -1.34248 1.34248 0.92807 -2.27054 1.34248 -0.01963
#> 9 10
#> 1.26385 -2.58670
#>
#> Coefficients: (1 not defined because of singularities)
#> Estimate Std. Error t value Pr(>|t|)
#> matstrawberry 8.9087 2.0267 4.396 0.00705 **
#> matpoppyseed 6.5427 1.5544 4.209 0.00842 **
#> matorange NA NA NA NA
#> matpear 1.2800 2.3056 0.555 0.60269
#> matmint 0.6582 2.6242 0.251 0.81193
#> matapple 1.2595 2.2526 0.559 0.60019
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 2.357 on 5 degrees of freedom
#> Multiple R-squared: 0.9504, Adjusted R-squared: 0.9007
#> F-statistic: 19.15 on 5 and 5 DF, p-value: 0.002824As you can see lm realizes that “1 [column] not defined
because of singularities” (matorange is not defined) but it
doesn’t indicate what columns it is linearly dependent with.
So if you would just look at the columns and not consider the
NA further, you would interpret that
strawberry and poppyseed are the biggest
contributors to the sweetness of fruit salads.
However, when you look at the model matrix you can see that the
orange column is a linear combination of the
strawberry and poppyseed columns (or vice
versa). So truly any of the three factors could contribute to the
sweetness of a fruit salad, the linear model has no way of recovering
which one given these 10 examples. And since we constructed this example
we know that orange and strawberry are the
sweetest and poppyseed contributes least to the
sweetness.
mat
#> strawberry poppyseed orange pear mint apple
#> [1,] 1 0 1 0 1 0
#> [2,] 1 0 1 0 1 0
#> [3,] 1 0 1 0 0 0
#> [4,] 1 0 1 1 0 0
#> [5,] 0 1 1 0 0 0
#> [6,] 0 1 1 0 0 0
#> [7,] 0 1 1 0 0 1
#> [8,] 0 0 0 1 0 1
#> [9,] 0 0 0 1 0 1
#> [10,] 0 0 0 1 0 1To make such cases more obvious and to be able to still correctly
interpret the linear model fit, we wrote fullRankMatrix. It
removes linearly dependent columns and renames the remaining columns to
make the dependencies clear using the
make_full_rank_matrix() function.
library(fullRankMatrix)
result <- make_full_rank_matrix(mat)
mat_fr <- result$matrix
space_list <- result$space_list
mat_fr
#> pear mint apple SPACE_1_AXIS1 SPACE_1_AXIS2
#> [1,] 0 1 0 -0.5 0.0000000
#> [2,] 0 1 0 -0.5 0.0000000
#> [3,] 0 0 0 -0.5 0.0000000
#> [4,] 1 0 0 -0.5 0.0000000
#> [5,] 0 0 0 0.0 -0.5773503
#> [6,] 0 0 0 0.0 -0.5773503
#> [7,] 0 0 1 0.0 -0.5773503
#> [8,] 1 0 1 0.0 0.0000000
#> [9,] 1 0 1 0.0 0.0000000
#> [10,] 1 0 1 0.0 0.0000000fit <- lm(sweetness ~ mat_fr + 0)
print(summary(fit))
#>
#> Call:
#> lm(formula = sweetness ~ mat_fr + 0)
#>
#> Residuals:
#> 1 2 3 4 5 6 7 8
#> -2.00934 2.00934 -1.34248 1.34248 0.92807 -2.27054 1.34248 -0.01963
#> 9 10
#> 1.26385 -2.58670
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> mat_frpear 1.2800 2.3056 0.555 0.60269
#> mat_frmint 0.6582 2.6242 0.251 0.81193
#> mat_frapple 1.2595 2.2526 0.559 0.60019
#> mat_frSPACE_1_AXIS1 -17.8174 4.0535 -4.396 0.00705 **
#> mat_frSPACE_1_AXIS2 -11.3322 2.6924 -4.209 0.00842 **
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 2.357 on 5 degrees of freedom
#> Multiple R-squared: 0.9504, Adjusted R-squared: 0.9007
#> F-statistic: 19.15 on 5 and 5 DF, p-value: 0.002824You can see that there are no more undefined columns. The columns
strawberry, orange and poppyseed
were removed and replaced with two columns (SPACE_1_AXIS1,
SPACE_1_AXIS2) that are linearly independent (orthogonal)
vectors that span the space in which the linearly dependent columns
strawberry, orange and poppyseed
lied.
The original columns that are contained within a space can be viewed
in the returned space_list:
space_list
#> $SPACE_1
#> [1] "strawberry" "poppyseed" "orange"In terms of interpretation the individual axes of the constructed
spaces are difficult to interpret, but we see that the axes of the space
of strawberry, orange and
poppyseed show a significant association with the sweetness
of fruit salads. A further resolution which of the three terms is most
strongly associated with sweetness is not possible with the
given number of observations, but there is definitely an association of
sweetness with the space spanned by the three terms.
If only a subset of all axes of a space show a significant association in the linear model fit, this could indicate that only a subset of linearly dependent columns that lie within the space spanned by the significantly associated axes drive this association. This would require some more detailed investigation by the user that would be specific to the use case.
There are already a few other packages out there that offer functions to detect linear dependent columns. Here are the ones we are aware of:
library(fullRankMatrix)
# let's say we have 10 fruit salads and indicate which ingredients are present in each salad
strawberry <- c(1,1,1,1,0,0,0,0,0,0)
poppyseed <- c(0,0,0,0,1,1,1,0,0,0)
orange <- c(1,1,1,1,1,1,1,0,0,0)
pear <- c(0,0,0,1,0,0,0,1,1,1)
mint <- c(1,1,0,0,0,0,0,0,0,0)
apple <- c(0,0,0,0,0,0,1,1,1,1)
# let's pretend we know how each fruit influences the sweetness of a fruit salad
# in this case we say that strawberries and oranges have the biggest influence on sweetness
set.seed(30)
strawberry_sweet <- strawberry * rnorm(10, 4)
poppyseed_sweet <- poppyseed * rnorm(10, 0.1)
orange_sweet <- orange * rnorm(10, 5)
pear_sweet <- pear * rnorm(10, 0.5)
mint_sweet <- mint * rnorm(10, 1)
apple_sweet <- apple * rnorm(10, 2)
sweetness <- strawberry_sweet + poppyseed_sweet+ orange_sweet + pear_sweet +
mint_sweet + apple_sweet
mat <- cbind(strawberry,poppyseed,orange,pear,mint,apple)caret::findLinearCombos(): https://rdrr.io/cran/caret/man/findLinearCombos.html
This function identifies which columns are linearly dependent and suggests which columns to remove. But it doesn’t provide appropriate naming for the remaining columns to indicate that any significant associations with the remaining columns are actually associations with the space spanned by the originally linearly dependent columns. Just removing the and then fitting the linear model would lead to erroneous interpretation.
caret_result <- caret::findLinearCombos(mat)Fitting a linear model with the orange column removed
would lead to the erroneous interpretation that strawberry
and poppyseed have the biggest influence on the fruit salad
sweetness, but we know it is actually
strawberry and orange.
mat_caret <- mat[, -caret_result$remove]
fit <- lm(sweetness ~ mat_caret + 0)
print(summary(fit))
#>
#> Call:
#> lm(formula = sweetness ~ mat_caret + 0)
#>
#> Residuals:
#> 1 2 3 4 5 6 7 8
#> -2.00934 2.00934 -1.34248 1.34248 0.92807 -2.27054 1.34248 -0.01963
#> 9 10
#> 1.26385 -2.58670
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> mat_caretstrawberry 8.9087 2.0267 4.396 0.00705 **
#> mat_caretpoppyseed 6.5427 1.5544 4.209 0.00842 **
#> mat_caretpear 1.2800 2.3056 0.555 0.60269
#> mat_caretmint 0.6582 2.6242 0.251 0.81193
#> mat_caretapple 1.2595 2.2526 0.559 0.60019
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 2.357 on 5 degrees of freedom
#> Multiple R-squared: 0.9504, Adjusted R-squared: 0.9007
#> F-statistic: 19.15 on 5 and 5 DF, p-value: 0.002824WeightIt::make_full_rank(): https://rdrr.io/cran/WeightIt/man/make_full_rank.html
This function removes some of the linearly dependent columns to create a full rank matrix, but doesn’t rename the remaining columns accordingly. For the user it isn’t clear which columns were linearly dependent and they can’t choose which column will be removed.
mat_weightit <- WeightIt::make_full_rank(mat, with.intercept = FALSE)
mat_weightit
#> strawberry poppyseed pear mint apple
#> [1,] 1 0 0 1 0
#> [2,] 1 0 0 1 0
#> [3,] 1 0 0 0 0
#> [4,] 1 0 1 0 0
#> [5,] 0 1 0 0 0
#> [6,] 0 1 0 0 0
#> [7,] 0 1 0 0 1
#> [8,] 0 0 1 0 1
#> [9,] 0 0 1 0 1
#> [10,] 0 0 1 0 1As above fitting a linear model with this full rank matrix would lead
to erroneous interpretation that strawberry and
poppyseed influence the sweetness, but we know
it is actually strawberry and orange.
fit <- lm(sweetness ~ mat_weightit + 0)
print(summary(fit))
#>
#> Call:
#> lm(formula = sweetness ~ mat_weightit + 0)
#>
#> Residuals:
#> 1 2 3 4 5 6 7 8
#> -2.00934 2.00934 -1.34248 1.34248 0.92807 -2.27054 1.34248 -0.01963
#> 9 10
#> 1.26385 -2.58670
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> mat_weightitstrawberry 8.9087 2.0267 4.396 0.00705 **
#> mat_weightitpoppyseed 6.5427 1.5544 4.209 0.00842 **
#> mat_weightitpear 1.2800 2.3056 0.555 0.60269
#> mat_weightitmint 0.6582 2.6242 0.251 0.81193
#> mat_weightitapple 1.2595 2.2526 0.559 0.60019
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 2.357 on 5 degrees of freedom
#> Multiple R-squared: 0.9504, Adjusted R-squared: 0.9007
#> F-statistic: 19.15 on 5 and 5 DF, p-value: 0.002824plm::detect.lindep(): https://rdrr.io/cran/plm/man/detect.lindep.html
The function returns which columns are potentially linearly dependent.
plm::detect.lindep(mat)
#> [1] "Suspicious column number(s): 1, 2, 3"
#> [1] "Suspicious column name(s): strawberry, poppyseed, orange"However it doesn’t capture all cases. For example here
plm::detect.lindep() says there are no dependent columns,
while there are several:
c1 <- rbinom(10, 1, .4)
c2 <- 1-c1
c3 <- integer(10)
c4 <- c1
c5 <- 2*c2
c6 <- rbinom(10, 1, .8)
c7 <- c5+c6
mat_test <- as.matrix(data.frame(c1,c2,c3,c4,c5,c6,c7))
plm::detect.lindep(mat_test)
#> [1] "No linear dependent column(s) detected."fullRankMatrix captures these cases:
result <- make_full_rank_matrix(mat_test)
result$matrix
#> (c1_AND_c4) SPACE_1_AXIS1 SPACE_1_AXIS2
#> [1,] 1 0.0000000 4.111431e-16
#> [2,] 0 -0.4082483 -5.419613e-17
#> [3,] 1 0.0000000 7.071068e-01
#> [4,] 0 -0.4082483 1.083923e-17
#> [5,] 1 0.0000000 7.071068e-01
#> [6,] 0 -0.4082483 1.083923e-17
#> [7,] 0 -0.4082483 1.083923e-17
#> [8,] 0 -0.4082483 1.083923e-17
#> [9,] 1 0.0000000 0.000000e+00
#> [10,] 0 -0.4082483 1.083923e-17Smisc::findDepMat(): https://rdrr.io/cran/Smisc/man/findDepMat.html
NOTE: this package was removed from CRAN as of 2020-01-26 (https://CRAN.R-project.org/package=Smisc) due to failing checks.
This function indicates linearly dependent rows/columns, but it doesn’t state which rows/columns are linearly dependent with each other.
However, this function seems to not work well for one-hot encoded matrices and the package doesn’t seem to be updated anymore (s. this issue: https://github.com/pnnl/Smisc/issues/24).
# example provided by Smisc documentation
Y <- matrix(c(1, 3, 4,
2, 6, 8,
7, 2, 9,
4, 1, 7,
3.5, 1, 4.5), byrow = TRUE, ncol = 3)
Smisc::findDepMat(t(Y), rows = FALSE)Trying with the model matrix from our example above:
Smisc::findDepMat(mat, rows=FALSE)
#> Error in if (!depends[j]) { : missing value where TRUE/FALSE neededThese binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.