
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.

spot_funs()
worksThe goal of funspotr (R function spotter) is to make it easy to identify which R functions and packages are used in files and projects. It was initially written to create reference tables of the functions and packages used in a few popular github repositories1.
There are roughly three types of functions in funspotr:
list_files_*(): that identify files in a repository or
related locationspot_*(): that identify functions or packages in
filesfunspotr is set-up for parsing R, Rmarkdown or Quarto files. If you want to parse a Jupyter notebook you should first convert it to an appropriate file type. If you pass in a file type that is not recognized (e.g. a .txt file) funspotr will attempt to parse it as if it is a .R script.
funspotr is primarily designed for identifying the functions / packages in self-contained files or collections of self-contained files (e.g. a blogdown project2). Though see Package dependencies in another file for examples of using it in other contexts.
Install the latest stable version of funspotr from CRAN with:
install.packages("funspotr")You can install the development version of funspotr from GitHub with:
# install.packages("devtools")
devtools::install_github("brshallo/funspotr")funspotr can be used to create reference tables of the functions and packages used in R projects.
The primary function in funspotr is spot_funs() which
returns a dataframe showing the functions and associated packages used
in a file.
library(funspotr)
file_lines <- "
library(dplyr)
require(tidyr)
as_tibble(mpg) %>%
mutate(class = as.character(class)) %>%
group_by(class) %>%
nest() %>%
mutate(stats = purrr::map(data,
~lm(cty ~ hwy, data = .x)))
made_up_fun()
"
file_output <- tempfile(fileext = ".R")
writeLines(file_lines, file_output)
spot_funs(file_path = file_output)
#> # A tibble: 10 × 2
#> funs pkgs
#> <chr> <chr>
#> 1 library base
#> 2 require base
#> 3 as_tibble tidyr
#> 4 mutate dplyr
#> 5 as.character base
#> 6 group_by dplyr
#> 7 nest tidyr
#> 8 map purrr
#> 9 lm stats
#> 10 made_up_fun (unknown)funs: functions in filepkgs: best guess as to the package the functions came
fromfunspotr has a few list_files_*() functions that return
a dataframe of relative_paths and
absolute_paths of all the R, Rmarkdown, or quarto files in
a specified location (currently: github repo, gists, or local). These
can be combined with a variant of spot_funs() that maps the
function across each file path found,
spot_funs_files():
library(dplyr)
# repo for an old presentation I gave
gh_ex <- list_files_github_repo(
repo = "brshallo/feat-eng-lags-presentation",
branch = "main") %>%
spot_funs_files()
gh_ex
#> # A tibble: 4 × 3
#> relative_paths absolute_paths spotted
#> <chr> <chr> <list>
#> 1 R/Rmd-to-R.R https://raw.githubusercontent.com/… <named list>
#> 2 R/feat-engineering-lags.R https://raw.githubusercontent.com/… <named list>
#> 3 R/load-inspections-save-csv.R https://raw.githubusercontent.com/… <named list>
#> 4 R/types-of-splits.R https://raw.githubusercontent.com/… <named list>relative_paths : relative filepathabsolute_paths: absolute filepath (in this case URL to
raw file on github)spotted: purrr::safely() style list-column
of results4 from mapping
spot_funs() across absolute_paths.These results may then be unnested with the helper
funspotr::unnest_results() to provide a table of functions
and packages by filepath. This can be manipulated like any other
dataframe – say we want to filter to only those files where here, readr or rsample packages are
used.
gh_ex %>%
unnest_results() %>%
filter(pkgs %in% c("here", "readr", "rsample"))
#> # A tibble: 8 × 4
#> funs pkgs relative_paths absolute_paths
#> <chr> <chr> <chr> <chr>
#> 1 here here R/Rmd-to-R.R https://raw.githubus…
#> 2 read_csv readr R/feat-engineering-lags.R https://raw.githubus…
#> 3 initial_time_split rsample R/feat-engineering-lags.R https://raw.githubus…
#> 4 training rsample R/feat-engineering-lags.R https://raw.githubus…
#> 5 testing rsample R/feat-engineering-lags.R https://raw.githubus…
#> 6 sliding_period rsample R/feat-engineering-lags.R https://raw.githubus…
#> 7 write_csv readr R/load-inspections-save-csv.R https://raw.githubus…
#> 8 here here R/load-inspections-save-csv.R https://raw.githubus…The outputs from funspotr::unnest_results() can also be
passed into funspotr::network_plot() to build a network
visualization of the connections between functions/packages and files5.
You might only want to parse certain file types or a subset of the files in a repo.
preview_files <- list_files_github_repo(
repo = "brshallo/feat-eng-lags-presentation",
branch = "main")
preview_files
#> # A tibble: 4 × 2
#> relative_paths absolute_paths
#> <chr> <chr>
#> 1 R/Rmd-to-R.R https://raw.githubusercontent.com/brshallo/feat…
#> 2 R/feat-engineering-lags.R https://raw.githubusercontent.com/brshallo/feat…
#> 3 R/load-inspections-save-csv.R https://raw.githubusercontent.com/brshallo/feat…
#> 4 R/types-of-splits.R https://raw.githubusercontent.com/brshallo/feat…Say we only want to parse the “types-of-splits.R” and “Rmd-to-R.R” files.
preview_files %>%
filter(stringr::str_detect(relative_paths, "types-of-splits|Rmd-to-R")) %>%
spot_funs_files() %>%
unnest_results()
#> # A tibble: 24 × 4
#> funs pkgs relative_paths absolute_paths
#> <chr> <chr> <chr> <chr>
#> 1 purl knitr R/Rmd-to-R.R https://raw.githubusercontent.com/br…
#> 2 here here R/Rmd-to-R.R https://raw.githubusercontent.com/br…
#> 3 library base R/types-of-splits.R https://raw.githubusercontent.com/br…
#> 4 theme_set ggplot R/types-of-splits.R https://raw.githubusercontent.com/br…
#> 5 theme_bw ggplot R/types-of-splits.R https://raw.githubusercontent.com/br…
#> 6 set.seed base R/types-of-splits.R https://raw.githubusercontent.com/br…
#> 7 tibble dplyr R/types-of-splits.R https://raw.githubusercontent.com/br…
#> 8 rep base R/types-of-splits.R https://raw.githubusercontent.com/br…
#> 9 today lubridate R/types-of-splits.R https://raw.githubusercontent.com/br…
#> 10 days lubridate R/types-of-splits.R https://raw.githubusercontent.com/br…
#> # ℹ 14 more rowsNote that if you have a lot of files in a repo you may need to set-up sleep periods or clone the repo locally and then parse the files from there so as to stay within the limits of github API hits.
Functions created in the file as well as functions from unavailable
packages (or packages that don’t exist) will output as
pkgs = "(unknown)".
file_lines_missing_pkgs <- "
library(dplyr)
as_tibble(mpg)
hello_world <- function() print('hello world')
madeuppkg::made_up_fun()
hello_world()
"
missing_pkgs_ex <- tempfile(fileext = ".R")
writeLines(file_lines_missing_pkgs, missing_pkgs_ex)
spot_funs(file_path = missing_pkgs_ex)
#> # A tibble: 5 × 2
#> funs pkgs
#> <chr> <chr>
#> 1 library base
#> 2 as_tibble dplyr
#> 3 print base
#> 4 made_up_fun (unknown)
#> 5 hello_world (unknown)To spot which package a function is from you must have the
package installed locally. Hence for files on others’ github repos
or that you created on a different machine, it is a good idea to start
with funspotr::check_pkgs_availability() to see which
packages you are missing and install the missing packages locally. If
you don’t want to edit your global library you may want to use renv or other environment
management tools.
funspotr has an internal helper
funspotr::install_missing_pkgs() for installing missing
packages:
spot_pkgs(file_output) %>%
check_pkgs_availability() %>%
funspotr::install_missing_pkgs()Alternatively, you may want to clone the repository locally and then
use renv::dependencies() and only then start using
funspotr6.
spot_funs() is currently set-up for self-contained
files. But spot_funs_custom() allows the user to explicitly
specify pkgs where functions may come from. This is useful
in cases where the packages loaded are not in the same location as the
file_path (e.g. they are loaded via source(),
or a DESCRIPTION file, or some other workflow). For example, below is a
made-up example where the library() calls are made in a
separate file and source()’d in.
# file where packages are loaded
file_libs <- "library(dplyr)
library(lubridate)"
file_libs_output <- tempfile(fileext = ".R")
writeLines(file_libs, file_libs_output)
# File of interest where things happen
file_run <- glue::glue(
"source('{ file_libs_output }')
tibble::tibble(days_from_today = 0:10) %>%
mutate(date = today() + days(days_from_today))
",
file_libs_output = stringr::str_replace_all(file_libs_output, "\\\\", "/")
)
file_run_output <- tempfile(fileext = ".R")
writeLines(file_run, file_run_output)
# Identify packages using both files and then pass in explicitly to `spot_funs_custom()`
pkgs <- c(spot_pkgs(file_libs_output),
spot_pkgs(file_run_output, show_explicit_funs = TRUE))
spot_funs_custom(
pkgs = pkgs,
file_path = file_run_output)
#> # A tibble: 5 × 2
#> funs pkgs
#> <chr> <chr>
#> 1 source base
#> 2 tibble tibble
#> 3 mutate dplyr
#> 4 today lubridate
#> 5 days lubridateAlso see funspotr::spot_pkgs_from_description().
Passing in show_each_use = TRUE to ... in
spot_funs() or spot_funs_files() will return
all instances of a function call rather than just once for each
file.
Compared to the initial example, mutate() now shows-up
at both rows 4 and 8:
spot_funs(file_path = file_output, show_each_use = TRUE)
#> # A tibble: 11 × 2
#> funs pkgs
#> <chr> <chr>
#> 1 library base
#> 2 require base
#> 3 as_tibble tidyr
#> 4 mutate dplyr
#> 5 as.character base
#> 6 group_by dplyr
#> 7 nest tidyr
#> 8 mutate dplyr
#> 9 map purrr
#> 10 lm stats
#> 11 made_up_fun (unknown)To automatically have your packages used as the tags for a blog
post you can add an inline function
funspotr::spot_tags() to a bullet in the tags
or categories argument of your YAML header. For
example:
---
title: This is a post
author: brshallo
date: '2022-02-11'
tags: ["`r funspotr::spot_tags()`"]
slug: this-is-a-post
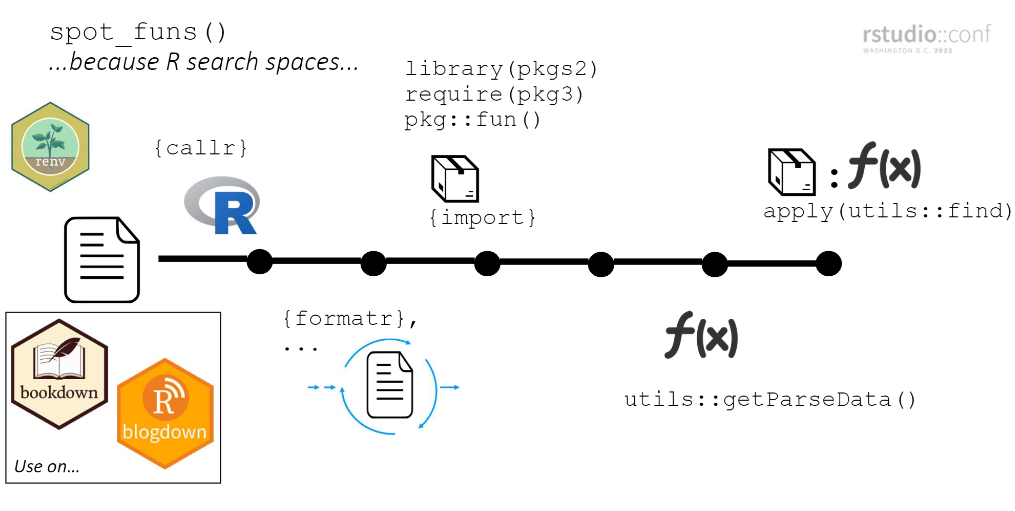
---spot_funs() worksfunspotr mimics the search space of each file prior to identifying
pkgs/funs. At a high-level…
pkg::fun()) are
loaded individually via import and are loaded
last (putting them at the top of the search space)7.(steps 1 and 2 needed so that step 4 has the best chance of identifying the package a function comes from in the file.)
utils::getParseData() and filter to
just functionsutils::find() to identify
associated packageExplainer slide from Rstudio Conf 2022 presentation:
mean in this
example: lapply(x, mean) . Similarly it will not identify
functions within switch(). See #13.knitr::read_chunk() and knitr::purl() in a
file passed to funspotr will also frequently cause an error in parsing.
See knitr#1753
& knitr#1938spot_funs() is primarily for cases where package
dependencies are loaded in the same file that they are used in10. Scripts that are not
self-contained typically should have the pkgs argument
provided explicitly via spot_funs_custom().spot_funs() through other
folder structures not yet mentioned.renv::dependencies() or a parsing
based approach. The simple regex’s I use have a variety of problems12.
R CMD check does function parsing. funspotr’s current
approach is comparatively slow and uses imperfect heuristics.+list_files*())list_files_github_repo() it may
make sense to instead clone the repo locally and then run
list_files_wd() from the repo prior to running
spot_funs_files() as this will limit the number of API hits
to github.Sys.sleep(), …).Prior posts (some of which used a now
deprecated API):
- Identifying
R Functions & Packages Used in GitHub Repos (funspotr part
1)
- Identifying
R Functions & Packages in Github Gists (funspotr part 2)
- Network
Plots of Code Collections (funspotr part 3)↩︎
Rather than, for example, targets workflows. Also, in some cases funspotr may not identify every function and/or package in a file (see Limitations, problems, musings or read the source code for details).↩︎
Other arguments may produce
additional columns. See spot_funs() reference page for
details.↩︎
list-column output where each item is
a list containing result and error.↩︎
renv is a more robust approach to finding and installing dependencies – particularly in cases where you are missing many dependencies or don’t want to alter the packages in your global library.↩︎
This heuristic is imperfect and means
that a file with “library(dplyr); select(); MASS::select()” would view
both select() calls as coming from {MASS} – when what it
should do is view the first was as coming from {dplyr} and the second
from {MASS}.↩︎
For example… If you pass in a .Rmd or .qmd that has a mix of R and python code chunks, the python chunks will simply be commented out. If you pass in a python script, you will almost certainly get a parsing error for that file.↩︎
In a language like python, where
calls are more explicit (e.g. np.*), all of the stuff with
recreating the search space would likely be unnecessary and you could
more easily just identify packages/functions by parsing the text.↩︎
i.e. in interactive R scripts or Rmd
or qmd documents where you use library() or related calls
within the script.↩︎
For example when reviewing David
Robinson’s Tidy Tuesday code I found that the meme package was used
far more than I would have expected. Turns out it was just due to it
reexporting the aes() function from ggplot.↩︎
e.g. in this case
lines <- "library(pkg)" the pkg would
show-up as a dependency despite just being part of a quote rather than
actually loaded. See #14 for
disucssion of other approaches.↩︎
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.