The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.

The goal of gghdx is to make it as simple as possible to follow the HDX visual guidelines when creating graphs using ggplot2. While most of the functionality is in allowing easy application of the HDX color ramps, the package also streamlines some of the other recommendations and best practices regarding plotted text, axis gridlines, and other visual features. The key functionalities are:
theme_hdx() is the general package theme.scale_color_hdx_...() and
scale_fill_hdx_...() applies the HDX color scale to the
relevant aesthetics.hdx_colors(), hdx_hex(), and
hdx_pal_...() provide easy user access to the HDX color
template. You can view the colorsgeom_text_hdx() and geom_label_hdx() wrap
the respective base functions to plot text using HDX fonts and
aesthetics.scale_y_continuous_hdx() wraps
scale_y_continuous() to plot data directly starting from
the y-axis.gghdx() ensures plot for the session use HDX defaults
for color and fill scales, uses theme_hdx() for all plots,
and applies scale_color_hdx_...() and
scale_fill_hdx_...()label_number_hdx() and format_number_hdx()
supplement the scales::label_...() series of functions to
format and create labels for numbers in the HDX style.You can install gghdx directly from CRAN:
install.packages("gghdx")You can install the development version from GitHub:
## install.packages("remotes")
remotes::install_github("OCHA-DAP/gghdx")The package is designed so the user just has to run
gghdx() once a session and mainly forget about it. This
will automatically set your ggplot2 to use the HDX theme, palettes,
fonts, and more by default. If you want more control or want to better
understand how the package works, please see the details below!
A quick and simple example would be plotting the iris
dataset included in base R.
library(ggplot2)
p <- ggplot(
iris,
aes(
x = Sepal.Length,
y = Petal.Length,
color = Species
)
) +
geom_point() +
labs(
title = "Iris species distributed by sepal and petal lengths",
y = "Petal length",
x = "Sepal length"
)
p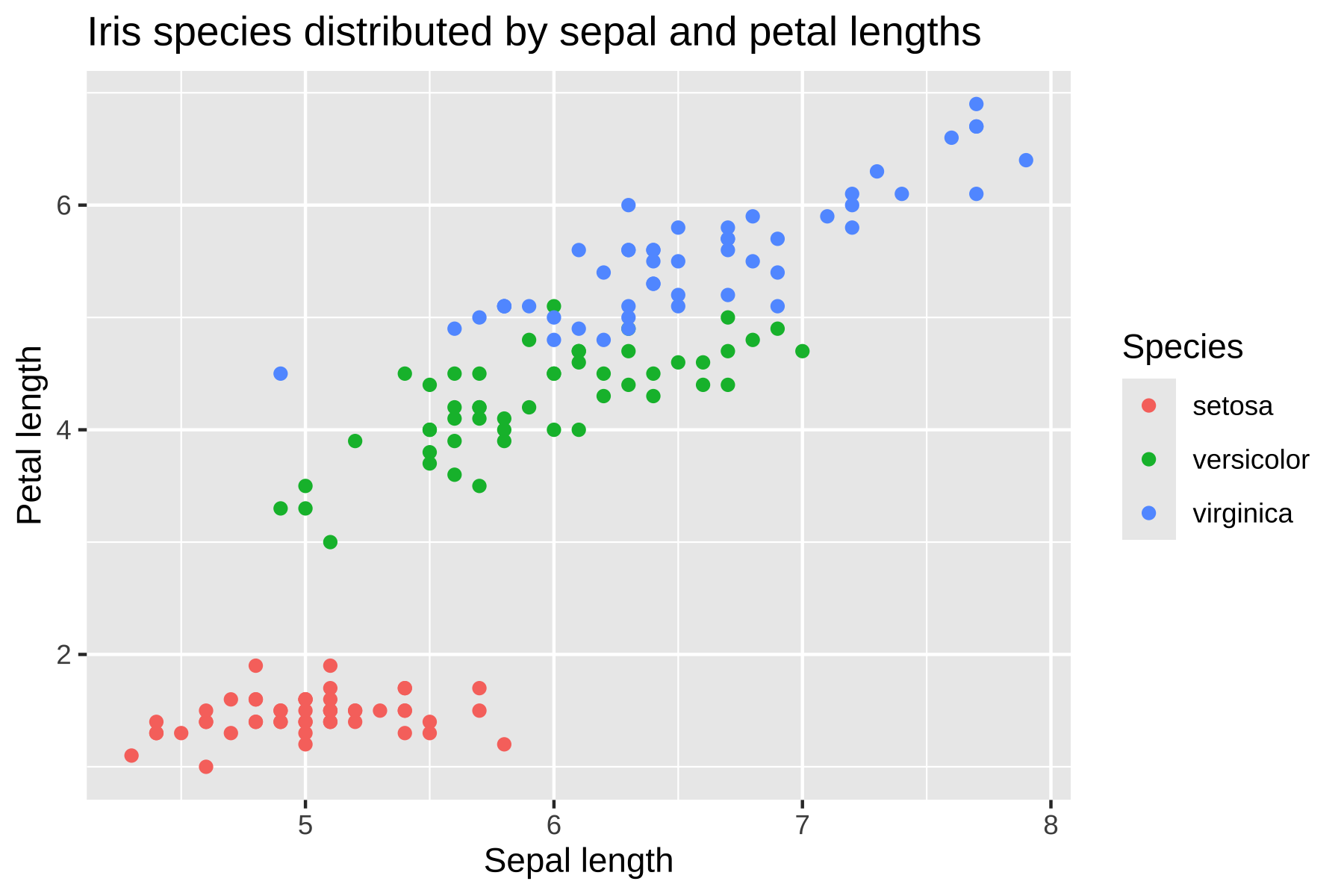
This output using the base ggplot style doesn’t look particularly
bad, but we can use theme_hdx() to quickly adjust some of
the styling to fit the style guide.
library(gghdx)
p + theme_hdx(base_family = "sans")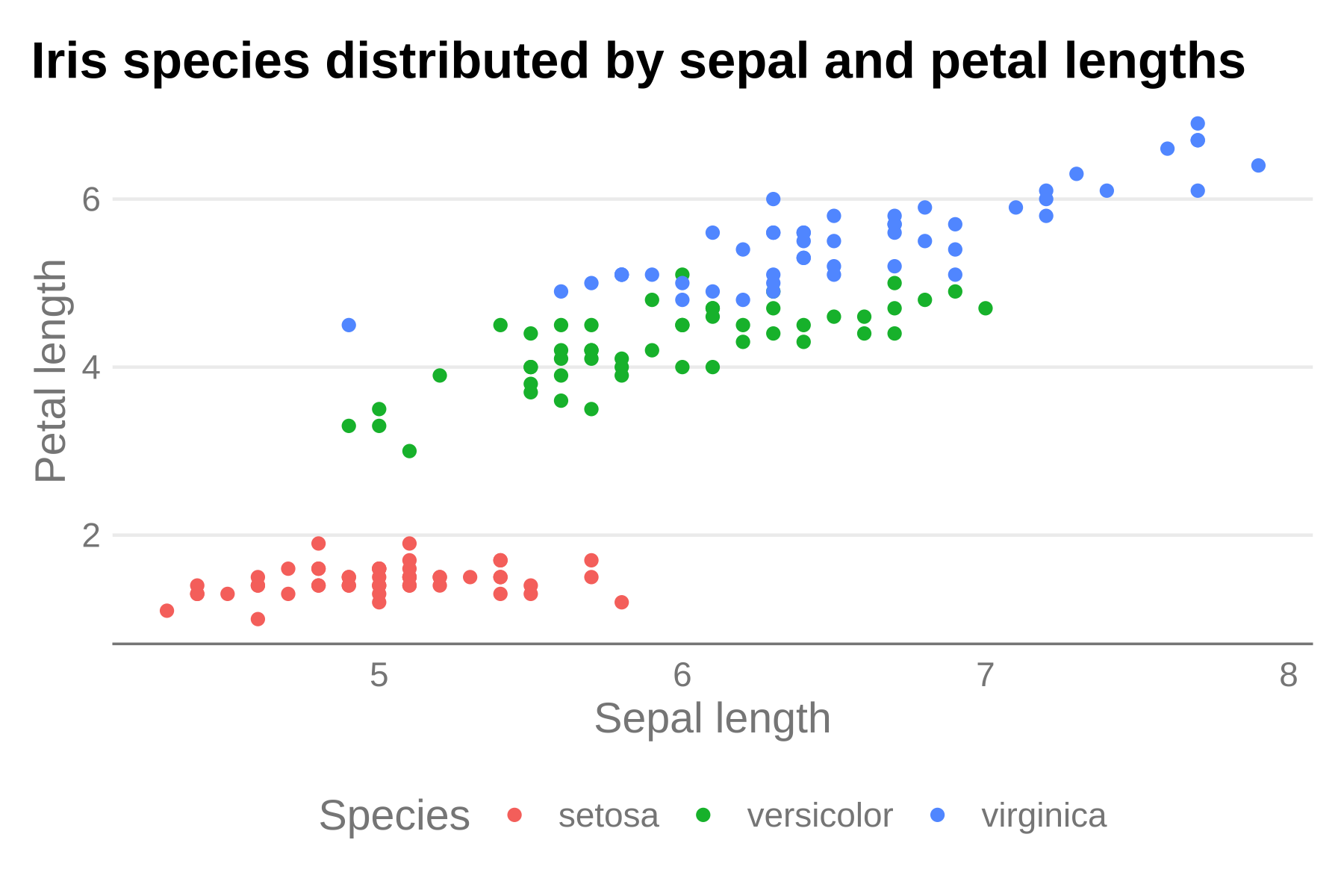
Now, axis lines have been cleaned up and the plot better resembles recommendations from the visual guide with just that single line of code.
However, the color palette for the points is still using the base R
palette. We can use one of the many scale_...hdx()
functions to use HDX colors. Let’s just use the primary discrete color
scale that will align each species with one of the 3 non-gray colorramps
(sapphire, mint, and tomato).
p + theme_hdx(base_family = "sans") + scale_color_hdx_discrete()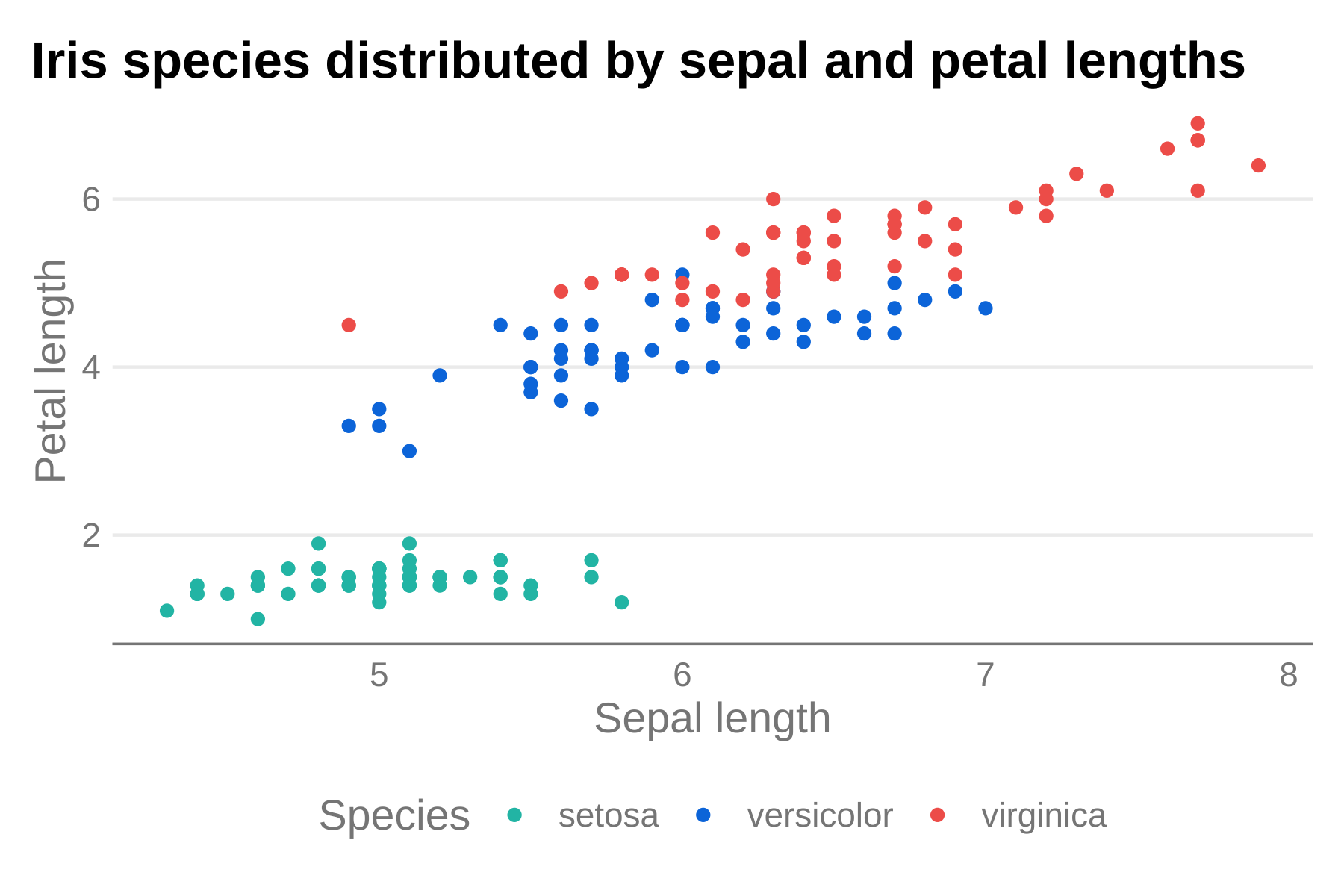
You can check the documentation of any of the
scale_...hdx() functions to see all available scales, or
directly access the colors using hdx_colors() or the raw
list in hdx_color_list. The available palettes can be
easily visualized using hdx_display_pal().
We also would like to use the HDX font family. Since Source Sans 3 is a free Google font, it makes it relatively easy to access in R. gghdx uses the sysfonts package to load the Google font and then showtext to include them in our plot. You can also use the extrafont package as an alternative if you have the font installed locally. This requires ghostscript to be installed locally and can run into other issues, such as font names not being found.
Below, I use the showtext package because it’s simpler.
library(showtext)
#> Loading required package: sysfonts
#> Loading required package: showtextdb
font_add_google("Source Sans 3")
showtext_auto()
p + theme_hdx(base_family = "Source Sans 3") + scale_color_hdx_discrete()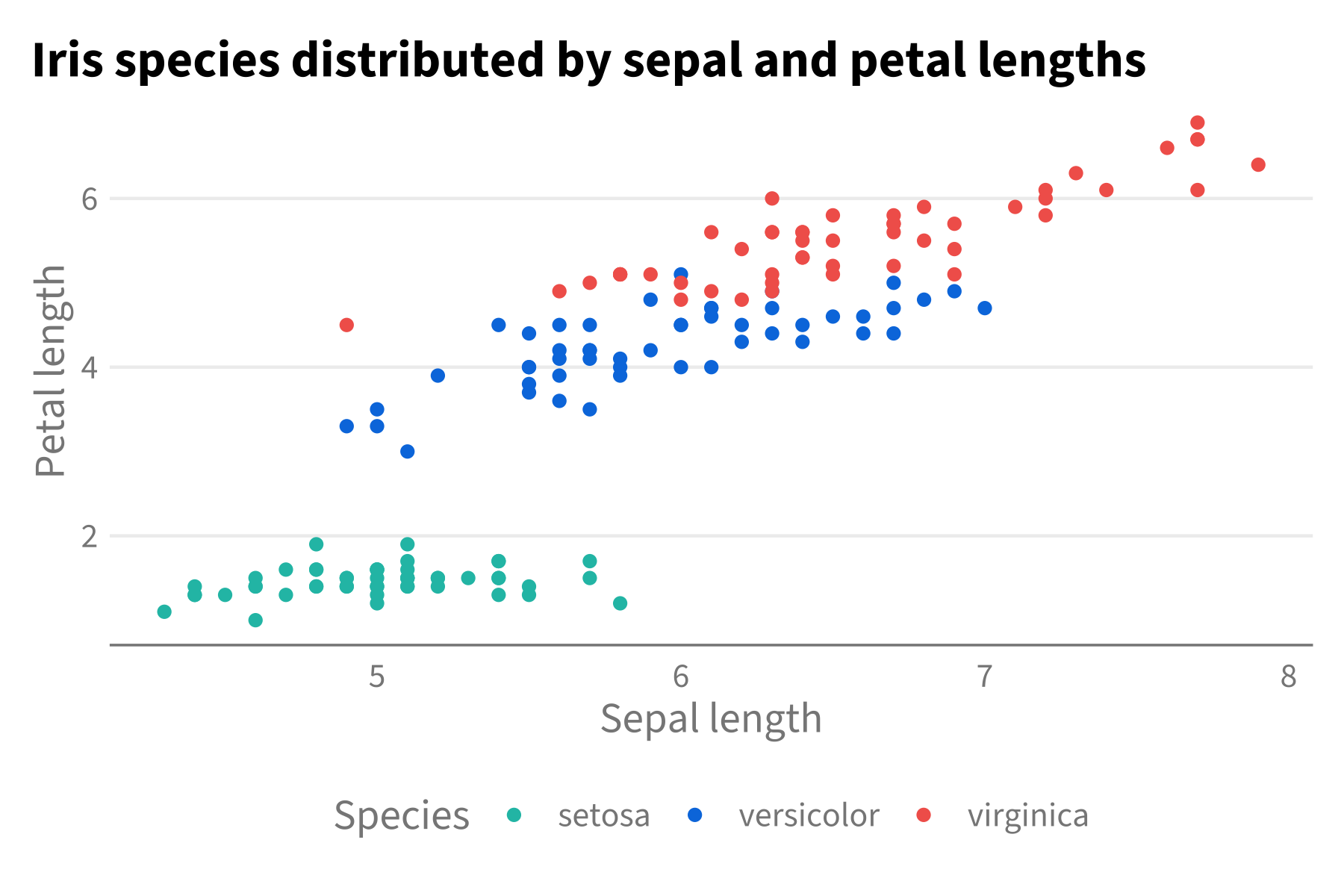
As clear above, even though we have an HDX theme function, we still
have to separately call the scale function to adjust our colors. And we
have to call these every time we make a new plot. So, to make life
simpler, gghdx() is provided as a convenience function that
sets ggplot to:
scale_fill_hdx_discrete() and
scale_color_hdx_discrete() as the default discrete fill and
color respectively;scale_fill_gradient_hdx_mint() and
scale_color_gradient_hdx_sapphire() as the default
continuous fill and color;You just have to run gghdx() once a session, and then
our plots will already be where we would like!
gghdx()
p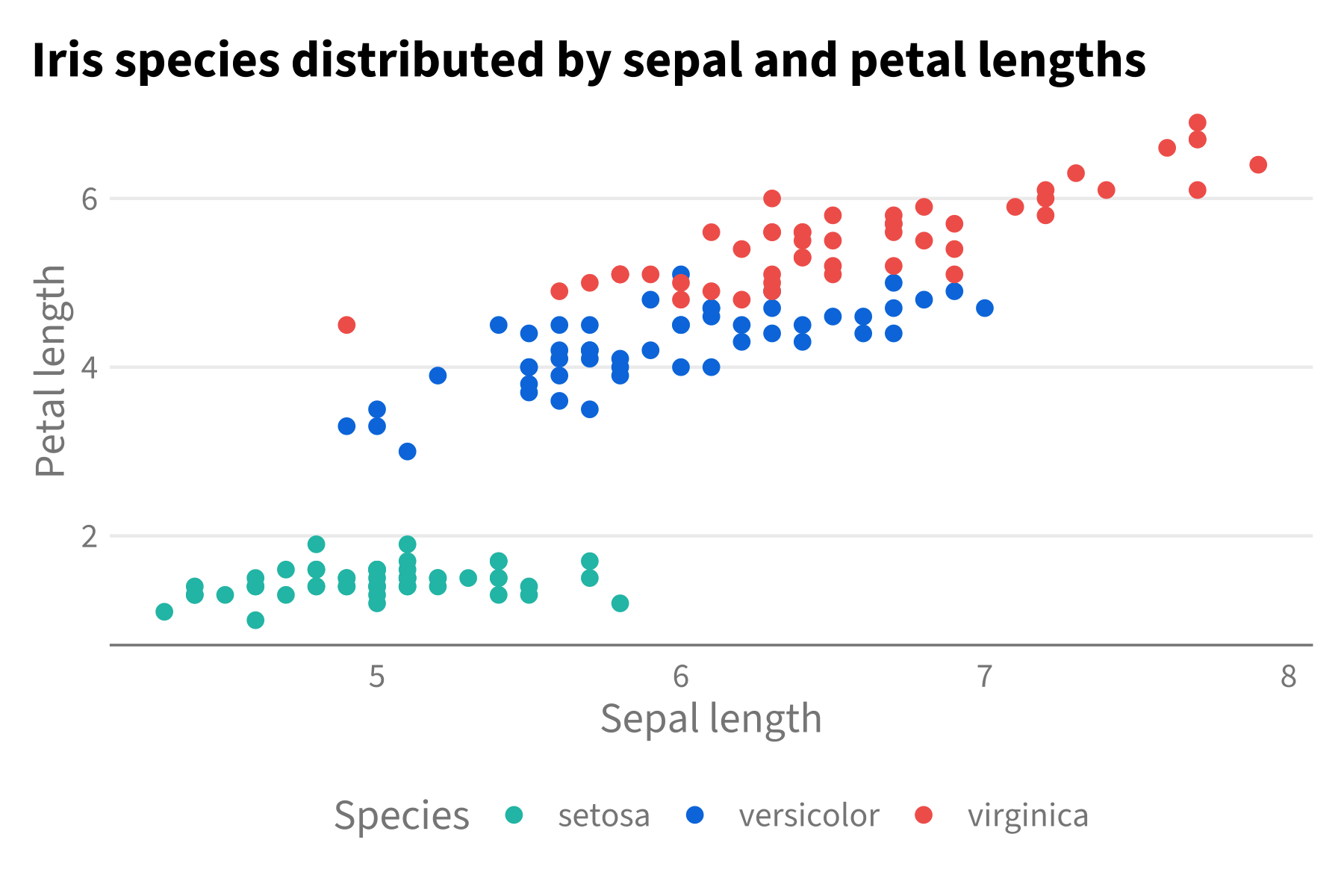
And voíla, we have our graph without specifying the theme or color scale.
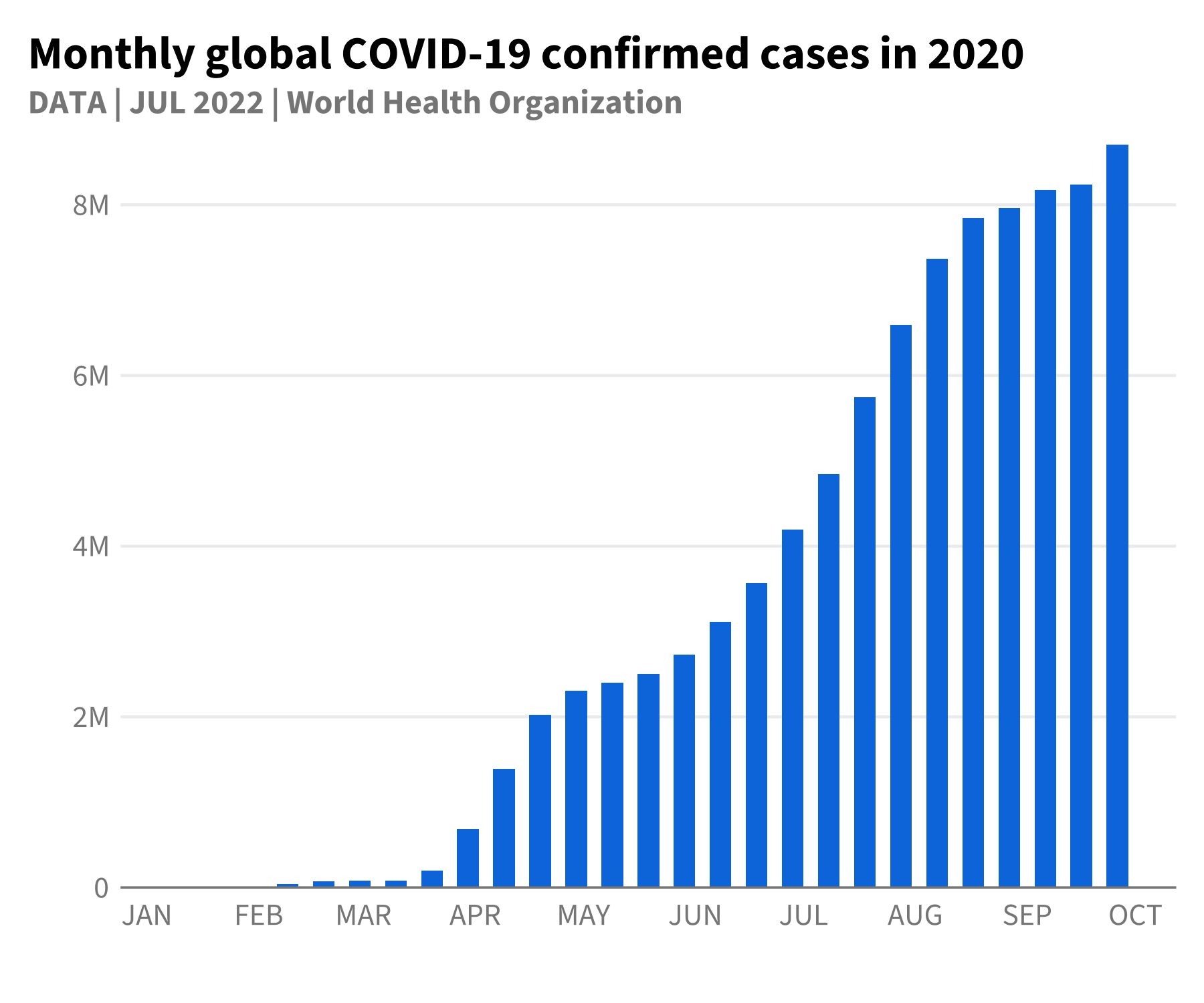
As a final example, we can closely match the COVID plots referenced in the visual guide using the theme and color scales in the package.
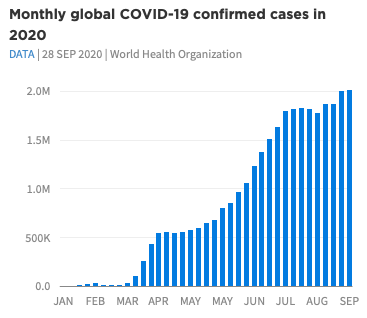
The inbuilt data gghdx::df_covid has aggregated COVID
data we can use to mirror this plot. To make the data start at the
y-axis, we can use scale_y_continuous_hdx() which sets
expand = c(0, 0) by default, and the
label_number_hdx() function to create custom labels.
p_blue <- ggplot(
df_covid,
aes(
x = date,
y = cases_monthly
)
) +
geom_bar(
stat = "identity",
width = 6,
fill = hdx_hex("sapphire-hdx") # use sapphire for fill
) +
scale_y_continuous_hdx(
labels = label_number_hdx()
) +
scale_x_date(
date_breaks = "1 month",
labels = function(x) toupper(strftime(x, "%b"))
) +
labs(
title = "Monthly global COVID-19 confirmed cases in 2020",
subtitle = "DATA | JUL 2022 | World Health Organization",
x = "",
y = ""
)
p_blue
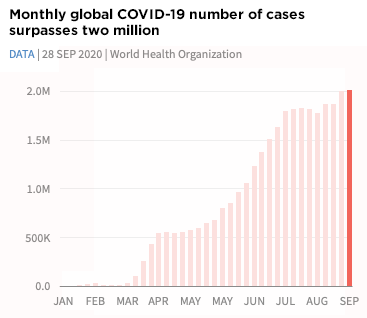
# create red plot
p_blue +
geom_bar(
aes(
fill = flag
),
width = 6,
stat = "identity"
) +
scale_fill_hdx_tomato() +
theme(
legend.position = "none"
) +
labs(
title = "Monthly COVID-19 # of cases surpasses 8 million"
)
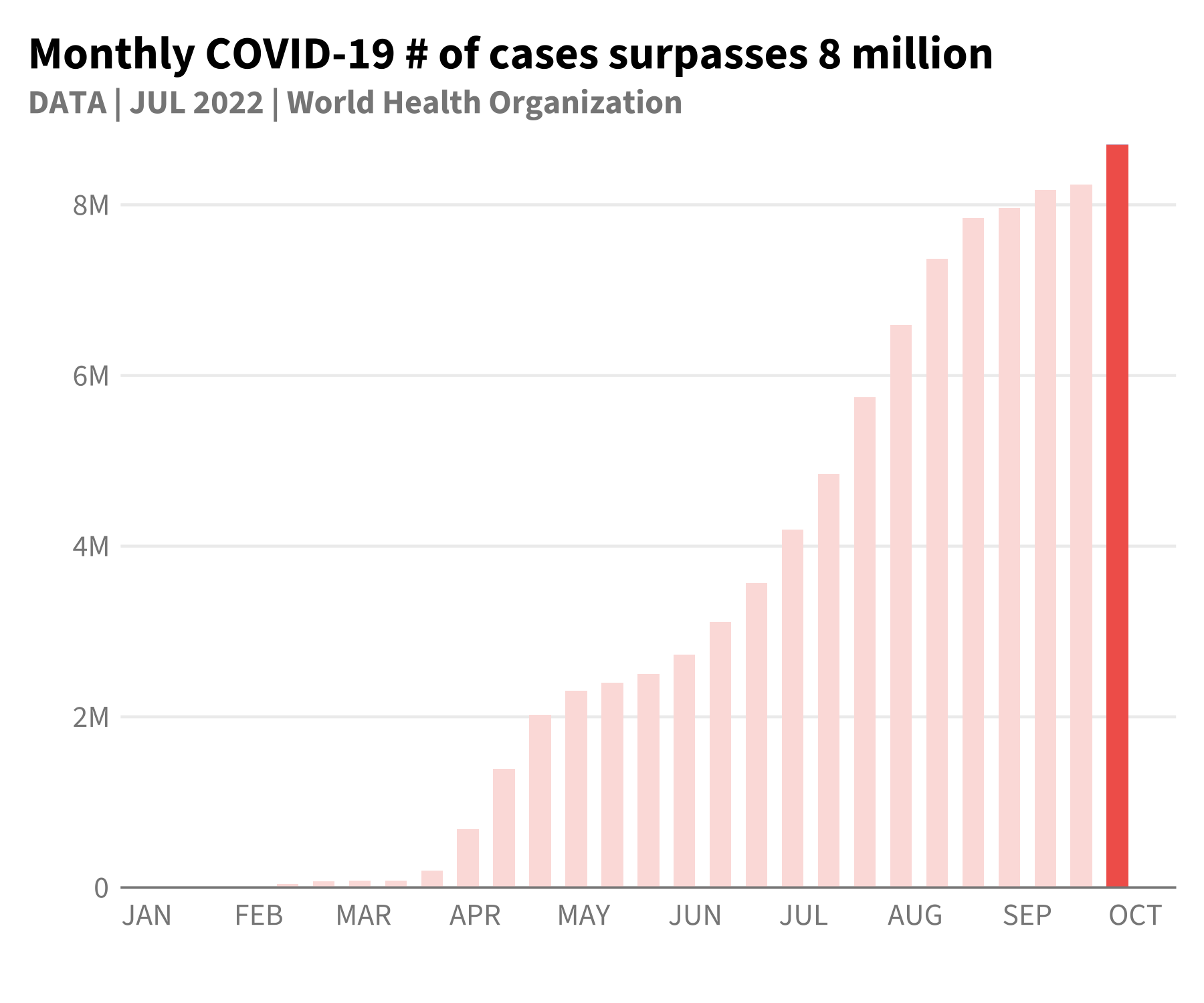
We’ve used relatively few lines of code to match fairly closely these examples plots!
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.