
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
ggseg.formats provides the ggseg_atlas S3 class that
powers the ggseg
ecosystem for 2D and 3D brain visualisation. It ships three bundled
atlases, a set of accessor functions for querying atlas contents, and a
pipe-friendly manipulation API for subsetting, renaming, and enriching
atlas objects.
The package includes three atlases covering the main atlas types:
library(ggseg.formats)plot(dk())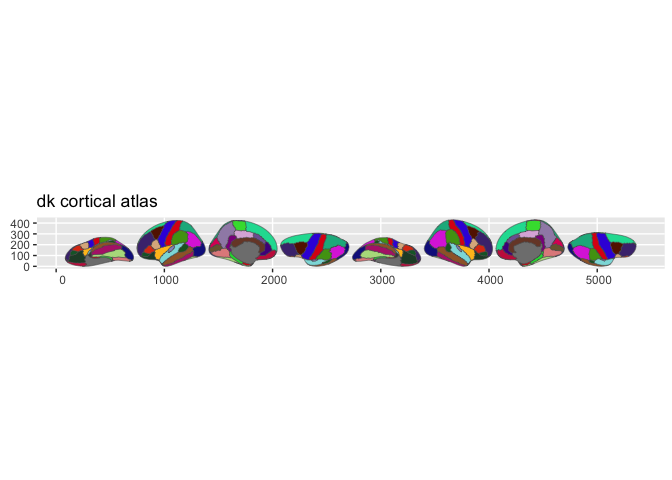
plot(aseg())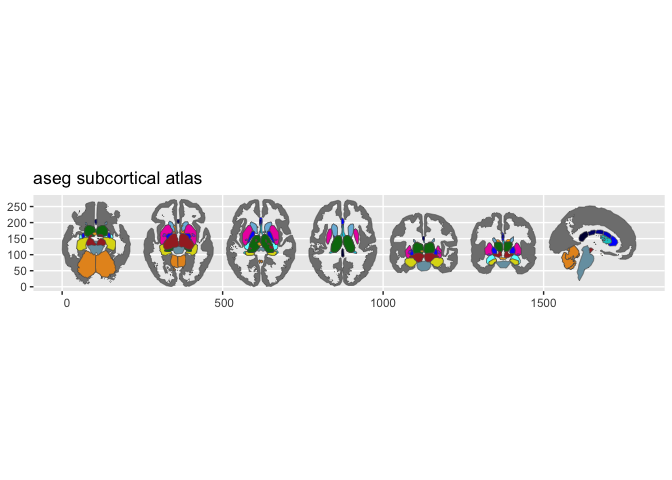
plot(tracula())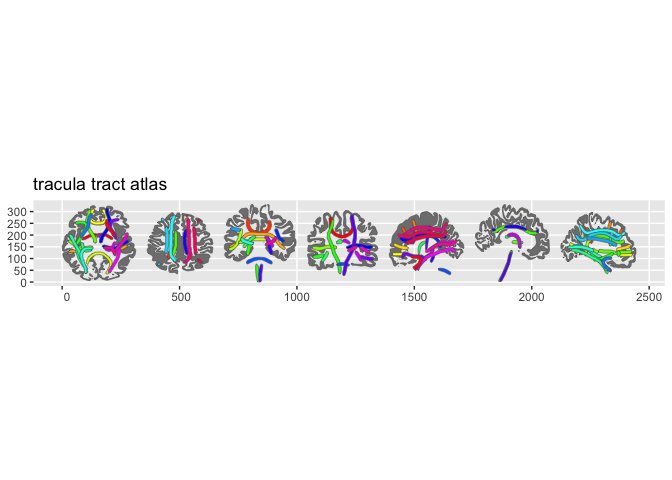
Atlas objects are designed for exploration and customisation. You can query regions, filter views, and pipe operations together:
aseg_small <- aseg() |>
atlas_region_keep("hippocampus|amygdala|thalamus") |>
atlas_view_keep("coronal_3|axial_3") |>
atlas_view_gather()
plot(aseg_small)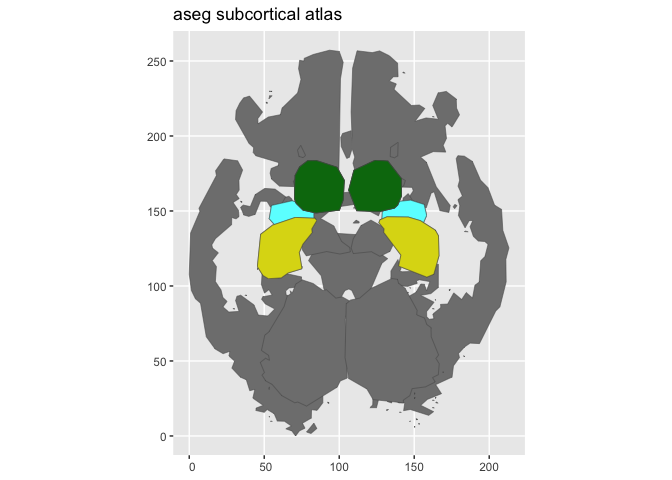
Install from the ggsegverse r-universe:
options(repos = c(
ggsegverse = 'https://ggsegverse.r-universe.dev',
CRAN = 'https://cloud.r-project.org'))
install.packages('ggseg.formats')Or install the development version from GitHub:
# install.packages("devtools")
devtools::install_github("ggseg/ggseg.formats")vignette("ggseg.formats") — understanding atlas
structure and accessorsvignette("atlas-manipulation") — region, view, and
metadata manipulationThese binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.