The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
Process discovery with variants of the Heuristics Miner algorithm
Discovery of process models from event logs based on the Heuristics Miner algorithm integrated into the bupaR framework.
You can install the release CRAN version with:
install.packages("heuristicsmineR")You can install the development version of heuristicsmineR with:
source("https://install-github.me/r-lib/remotes")
remotes::install_github("bupaverse/heuristicsmineR")This is a basic usage example discovering the Causal net of the
patients event log:
library(heuristicsmineR)
library(eventdataR)
data(patients)
# Dependency graph / matrix
dependency_matrix(patients)
# Causal graph / Heuristics net
causal_net(patients)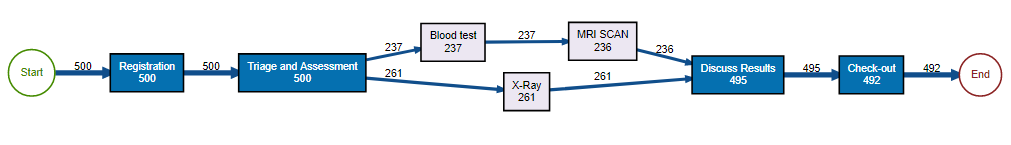
This discovers the Causal net of the built-in L_heur_1
event log that was proposed in the book Process
Mining: Data Science in Action:
# Efficient precedence matrix
m <- precedence_matrix_absolute(L_heur_1)
as.matrix(m)
# Example from Process mining book
dependency_matrix(L_heur_1, threshold = .7)
causal_net(L_heur_1, threshold = .7)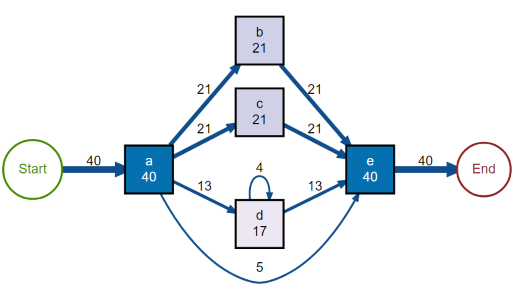
The Causal net can be converted to a Petri net (note that there are some unnecessary invisible transition that are not yet removed):
# Convert to Petri net
library(petrinetR)
cn <- causal_net(L_heur_1, threshold = .7)
pn <- as.petrinet(cn)
render_PN(pn)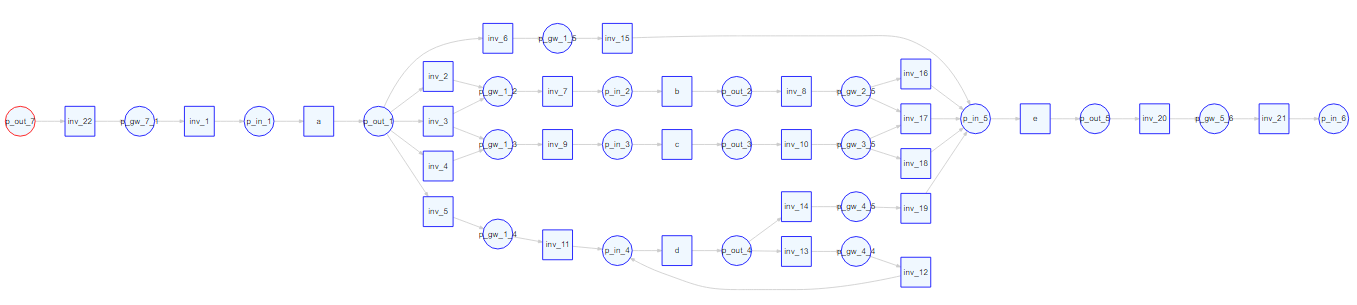
The Petri net can be further used, for example for conformance checking through the pm4py package (Note that the final marking is currently not saved in petrinetR):
library(pm4py)
conformance_alignment(L_heur_1, pn,
initial_marking = pn$marking,
final_marking = c("p_in_6"))These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.