The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
The goal of inlabru is to facilitate
spatial modeling using integrated nested Laplace approximation via the
R-INLA package. Additionally,
extends the GAM-like model class to more general nonlinear predictor
expressions, and implements a log Gaussian Cox process likelihood for
modeling univariate and spatial point processes based on ecological
survey data. Model components are specified with general inputs and
mapping methods to the latent variables, and the predictors are
specified via general R expressions, with separate expressions for each
observation likelihood model in multi-likelihood models. A prediction
method based on fast Monte Carlo sampling allows posterior prediction of
general expressions of the latent variables. See Fabian E. Bachl, Finn
Lindgren, David L. Borchers, and Janine B. Illian (2019), inlabru: an R
package for Bayesian spatial modelling from ecological survey data,
Methods in Ecology and Evolution, British Ecological Society, 10,
760–766, doi:10.1111/2041-210X.13168,
and citation("inlabru").
The inlabru.org website has links to old tutorials with code examples for versions up to 2.1.13. For later versions, updated versions of these tutorials, as well as new examples, can be found at https://inlabru-org.github.io/inlabru/articles/
You can install the current CRAN version version of inlabru:
options(repos = c(
INLA = "https://inla.r-inla-download.org/R/testing",
getOption("repos")
))
install.packages("inlabru")You can install the latest bugfix release of inlabru from GitHub with:
# install.packages("pak")
pak::repo_add(INLA = "https://inla.r-inla-download.org/R/testing")
pak::pkg_install("inlabru-org/inlabru@stable")You can install the development version of inlabru from GitHub with
pak::pkg_install("inlabru-org/inlabru")or track the development version builds via inlabru-org.r-universe.dev:
# Enable universe(s) by inlabru-org
pak::repo_add(inlabruorg = "https://inlabru-org.r-universe.dev")
pak::pkg_install("inlabru")This will pick the r-universe version if it is more recent than the CRAN version.
remotesYou can install the latest bugfix release of inlabru from GitHub with:
# install.packages("remotes")
remotes::install_github("inlabru-org/inlabru", ref = "stable")You can install the development version of inlabru from GitHub with
remotes::install_github("inlabru-org/inlabru", ref = "devel")or track the development version builds via inlabru-org.r-universe.dev:
# Enable universe(s) by inlabru-org
options(repos = c(
inlabruorg = "https://inlabru-org.r-universe.dev",
INLA = "https://inla.r-inla-download.org/R/testing",
CRAN = "https://cloud.r-project.org"
))
# Install some packages
install.packages("inlabru")This is a basic example which shows how fit a simple spatial Log Gaussian Cox Process (LGCP) and predicts its intensity:
# Load libraries
library(INLA)
#> Loading required package: Matrix
#> This is INLA_25.06.22-1 built 2025-06-22 16:17:48 UTC.
#> - See www.r-inla.org/contact-us for how to get help.
#> - List available models/likelihoods/etc with inla.list.models()
#> - Use inla.doc(<NAME>) to access documentation
library(inlabru)
#> Loading required package: fmesher
library(fmesher)
library(ggplot2)
# Construct latent model components
matern <- inla.spde2.pcmatern(
gorillas_sf$mesh,
prior.sigma = c(0.1, 0.01),
prior.range = c(0.01, 0.01)
)
cmp <- ~ mySmooth(geometry, model = matern) + Intercept(1)
# Fit LGCP model
# This particular bru/bru_obs combination has a shortcut function lgcp() as well
fit <- bru(
cmp,
bru_obs(
formula = geometry ~ .,
family = "cp",
data = gorillas_sf$nests,
samplers = gorillas_sf$boundary,
domain = list(geometry = gorillas_sf$mesh)
),
options = list(control.inla = list(int.strategy = "eb"))
)
# Predict Gorilla nest intensity
lambda <- predict(
fit,
fm_pixels(gorillas_sf$mesh, mask = gorillas_sf$boundary),
~ exp(mySmooth + Intercept)
)# Plot the result
ggplot() +
geom_fm(data = gorillas_sf$mesh) +
gg(lambda, geom = "tile") +
gg(gorillas_sf$nests, color = "red", size = 0.5, alpha = 0.5) +
ggtitle("Nest intensity per km squared") +
xlab("") +
ylab("")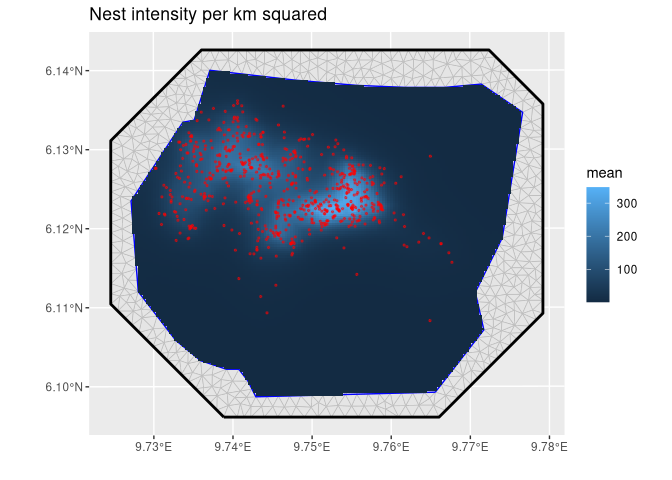
Nest intensity per km squared
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.