
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
The goal of lazytrade is to keep all functions and scripts of the lazytrade educational project on UDEMY. Functions are providing an opportunity to learn Computer and Data Science using example of Algorithmic Trading. Please kindly not that this project was created for Educational Purposes only!
You can install the released version of lazytrade from CRAN with:
install.packages("lazytrade")And the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("vzhomeexperiments/lazytrade")This is a basic example which shows you how to solve a common problem:
library(lazytrade)
library(magrittr, warn.conflicts = FALSE)
## basic example code
# Convert a time series vector to matrix with 64 columns
macd_m <- seq(1:1000) %>% as.data.frame() %>% to_m(20)
head(macd_m, 2)
#> [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10] [,11] [,12] [,13] [,14]
#> [1,] 1 2 3 4 5 6 7 8 9 10 11 12 13 14
#> [2,] 21 22 23 24 25 26 27 28 29 30 31 32 33 34
#> [,15] [,16] [,17] [,18] [,19] [,20]
#> [1,] 15 16 17 18 19 20
#> [2,] 35 36 37 38 39 40Why is it useful? It is possible to convert time-series data into matrix data to do make modeling
Multiple log files could be joined into one data object
library(lazytrade)
library(readr)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
library(magrittr)
library(lubridate)
#>
#> Attaching package: 'lubridate'
#> The following objects are masked from 'package:base':
#>
#> date, intersect, setdiff, union
# files are located in the sample folders
DFOLDER <- system.file("extdata/RES", package = "lazytrade")
DFR <- opt_aggregate_results(path_data = DFOLDER)This data object can be visualized
library(ggplot2)
opt_create_graphs(x = DFR, outp_path = dir,graph_type = 'bars')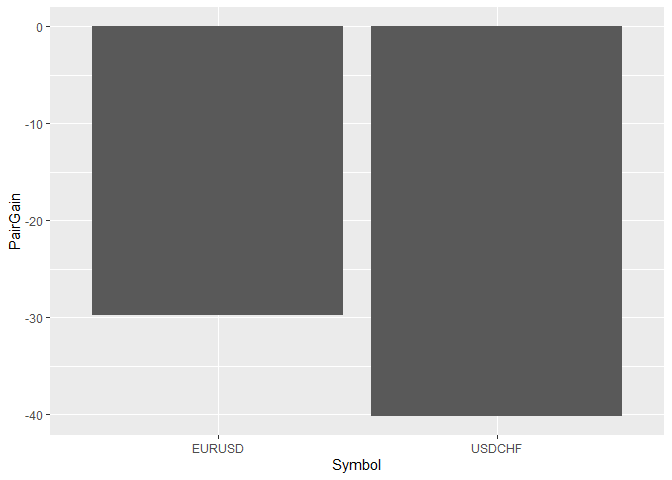
Or just visualize results with time-series plot
opt_create_graphs(x = DFR, outp_path = dir,graph_type = 'ts')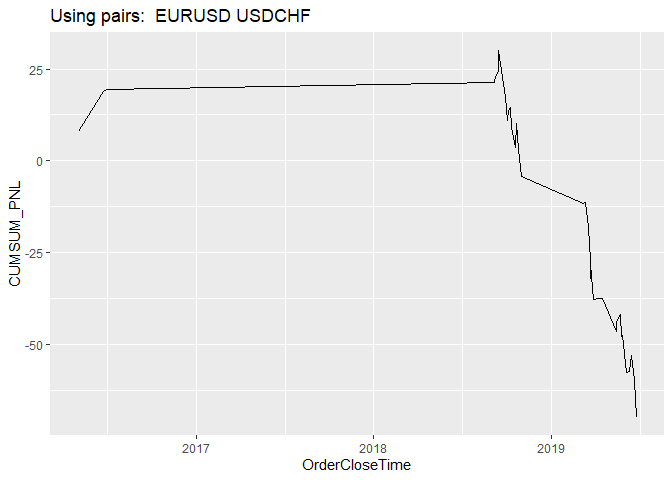
Example below would generate RL policy based on the trade results achieved so far
library(dplyr)
library(ReinforcementLearning)
library(magrittr)
data(data_trades)
states <- c("tradewin", "tradeloss")
actions <- c("ON", "OFF")
control <- list(alpha = 0.7, gamma = 0.3, epsilon = 0.1)
rl_generate_policy(data_trades, states, actions, control)
#> TradeState Policy
#> tradeloss tradeloss ON
#> tradewin tradewin OFFMultiple trading accounts require passwords, package contains function that may easily generate random passwords:
library(lazytrade)
library(stringr)
library(magrittr)
library(openssl)
library(readr)
#generate 8digit password for trading platform
util_generate_password(salt = 'random text')
#> .
#> 1 ac5cE049Facilitate generation of initialisation files:
library(lazytrade)
dir <- normalizePath(tempdir(),winslash = "/")
# test file to launch MT4 terminal with parameters
write_ini_file(mt4_Profile = "Default",
mt4_Login = "12345678",
mt4_Password = "password",
mt4_Server = "BrokerServerName",
dss_inifilepath = dir,
dss_inifilename = "prod_T1.ini",
dss_mode = "prod")What is special about using README.Rmd instead of just
README.md? You can include R chunks like so:
summary(cars)
#> speed dist
#> Min. : 4.0 Min. : 2.00
#> 1st Qu.:12.0 1st Qu.: 26.00
#> Median :15.0 Median : 36.00
#> Mean :15.4 Mean : 42.98
#> 3rd Qu.:19.0 3rd Qu.: 56.00
#> Max. :25.0 Max. :120.00You’ll still need to render README.Rmd regularly, to
keep README.md up-to-date.
taken from https://r-pkgs.org/
taken from https://lifecycle.r-lib.org/articles/communicate.html
Run Once:
usethis::use_lifecycle()
To insert badge:
Add badges in documentation topics by inserting one of:
#’ #’ #’
Create right title case for the title of the package By running this
command…
tools::toTitleCase("Learn computer and data science using algorithmic trading")
the Title will become: “Learn Computer and Data Science using
Algorithmic Trading”
Run this code to re-generate documentation
devtools::document()
Run this code to fix license:
usethis::use_mit_license(name = "Vladimir Zhbanko")
Run this code to add data to the folder data/
x <- sample(1000) usethis::use_data(x)
To update this data: x <- sample(2000)
usethis::use_data(x, overwrite = T)
To convert character into time:
mutate(across('X1', ~ as.POSIXct(.x, format = "%Y.%m.%d %H:%M:%S")))
Note: use option ’LazyLoad` to make data available only when user wants it always include LazyData: true in your DESCRIPTION. Note: to document dataset see https://stackoverflow.com/questions/2310409/how-can-i-document-data-sets-with-roxygen
Document dataset using the R script R/datasets.R
Use data in the function with data(x)
Place data like small files to the folder:
inst/extdata
Run this command to setup tests ‘usethis::use_testthat()’
This will create a folder with the name tests
Inside this folder there will be another folder
testthat.
@examples …
code to execute during package checks
@examples
/donttest{
code to NOT execute during package checks
}
Run this command to create a new script with the test skeleton:
usethis::use_test("profit_factor.R")
Details:
context("profit_factor")data(named_data_object)Example:
library(testthat)
#>
#> Attaching package: 'testthat'
#> The following object is masked from 'package:dplyr':
#>
#> matches
#> The following objects are masked from 'package:readr':
#>
#> edition_get, local_edition
#> The following objects are masked from 'package:magrittr':
#>
#> equals, is_less_than, not
library(dplyr)
library(magrittr)
context("profit_factor")
test_that("test value of the calculation", {
data(profit_factor_data)
DF_Stats <- profit_factor_data %>%
group_by(X1) %>%
summarise(PnL = sum(X5),
NumTrades = n(),
PrFact = util_profit_factor(X5)) %>%
select(PrFact) %>%
head(1) %>%
pull(PrFact) %>%
round(3)
expect_equal(DF_Stats, 0.68)
})
#> Test passed 🎊Test coverage shows you what you’ve tested devtools::test_coverage_file()
devtools::test_coverage_file()
This will add automatic test coverage badge to the readme file on
github usethis::use_coverage()
Step 1. devtools::document() Step 2.
devtools::run_examples() Step 3. Menu ‘Build’
Clean and Rebuild Step 4. ‘Check’
devtools::check()
This is now a default option
Whenever examples construct is used author of the package must insure that those examples are running. Such examples are those that would require longer test execution. To perform this test package needs to be checked with the following command:
devtools::check(run_dont_test = TRUE)
whenever a quick check is required:
devtools::check(run_dont_test = FALSE) ???
In case functions are writing files there are few considerations to take into account:
tempdir() functiondevtools::check()
there should nothing remain in the ‘tmp/’ directoryFile names defined by function tempdir() would look like
this:
# > tempdir()
# [1] "/tmp/RtmpkaFStZ"File names defined by function tempfile() would look
like this:
# > tempfile()
# [1] "/tmp/RtmpkaFStZ/file7a33be992b4"This is example of how function write_csv example
works:
tmp <- tempfile()
write_csv(mtcars, tmp)results of this code are correctly stored to the temporary file
however this example from readr package function
write_csv is showing that file will be written to the
‘/tmp/’ directory
dir <- tempdir()
write_tsv(mtcars, file.path(dir, "mtcars.tsv.gz"))We use function unlink() to do this:
unlink("/tmp/*.csv", recursive = TRUE, force = TRUE)and we check that there is nothing more remained:
dir("/tmp/*.csv")To remove function from the package we can use:
see https://stackoverflow.com/questions/9439256/how-can-i-handle-r-cmd-check-no-visible-binding-for-global-variable-notes-when see https://github.com/HughParsonage/grattan/blob/master/R/zzz.R
After first submission there are some notes on specific R flavors
This question was addressed here but yet it’s not answered: https://stackoverflow.com/questions/48487541/r-cmd-check-note-namespace-in-imports-field-not-imported
To search for specific function in the scripts one can do the following:
list_of_functions <- c(
"drop_na",
"fill",
"extract",
"gather",
"nest",
"separate"
)
for (FN in list_of_functions) {
if(!exists("res")){
res <- BurStMisc::scriptSearch(FN)
} else {
res2 <- BurStMisc::scriptSearch(FN)
res <- mapply(c, res, res2, SIMPLIFY=FALSE)}
}It’s important to avoid that function write to the directory other
then tempdir() Construct file name must be done using
file.name() function as follow:
# use plane temp directory
dir_name <- normalizePath(tempdir(),winslash = "/")
file_name <- paste0('my_file', 1, '.csv')
# this needs to be used in the function
full_path <- file.path(dir_name, file_name)
# when using sub-directory
sub_dir <- file.path(dir_name, "_SUB")
if(!dir.exists(sub_dir)){dir.create(sub_dir)}https://r-pkgs.org/description.html#version
Clone package from GitHub and test check it in Docker Container
devtools::build()
usethis::use_readme_rmd()
usethis::use_github_action()
To be elaborated
Setup the new version of the package:
usethis::use_release_issue()
Follow checklist before upload to CRAN:
devtools::release_checks()
then:
devtools::release()
spelling devtools::spell_check()
See ?rhubv2
devtools::check_win_release()
devtools::check_win_devel()
devtools::check_win_oldrelease()
Explain the changes
https://cran.r-project.org/submit.html
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.