The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
The goal of lfproQC R package is to provide an optimal combination of normalization and imputation methods for the label-free proteomics expression dataset.
You can install the development version of lfproQC from GitHub with:
# install.packages("devtools")
devtools::install_github("kabilansbio/lfproQC", build_manual = TRUE, build_vignette = TRUE)This is a basic example for finding the best combinations of normalization and imputation method for the label-free proteomics expression dataset:
library(lfproQC)
## basic example code with the example dataset and data groups
yeast <- best_combination(yeast_data, yeast_groups, data_type = "Protein")
yeast$`Best combinations`
#> PCV_best_combination PEV_best_combination PMAD_best_combination
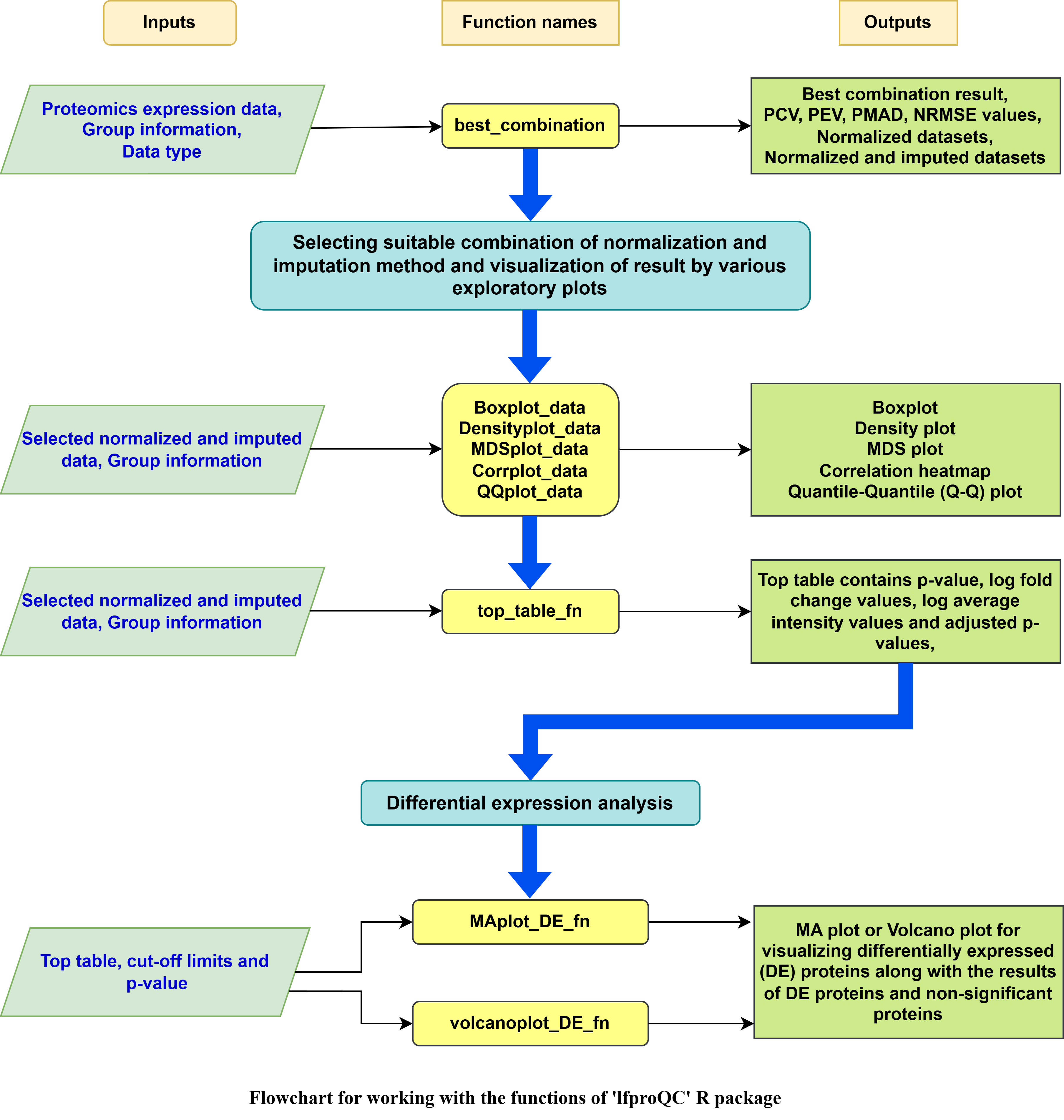
#> 1 rlr_knn, rlr_lls vsn_lls rlr_llsThe overall workflow for using the ‘lfproQC’ package

These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.