The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
An R package for generating concentration-time profiles from linear pharmacokinetic (PK) systems.
To install from CRAN:
install.packages("linpk")To install the latest development version directly from GitHub:
require(devtools)
devtools::install_github("benjaminrich/linpk")To simulate a PK profile from a one-compartment model with first-order absorption under repeated dosing:
t.obs <- seq(0, 6*24, 0.5)
y <- pkprofile(t.obs, cl=0.5, vc=11, ka=1.3, dose=list(amt=100, addl=9, ii=12))
plot(y)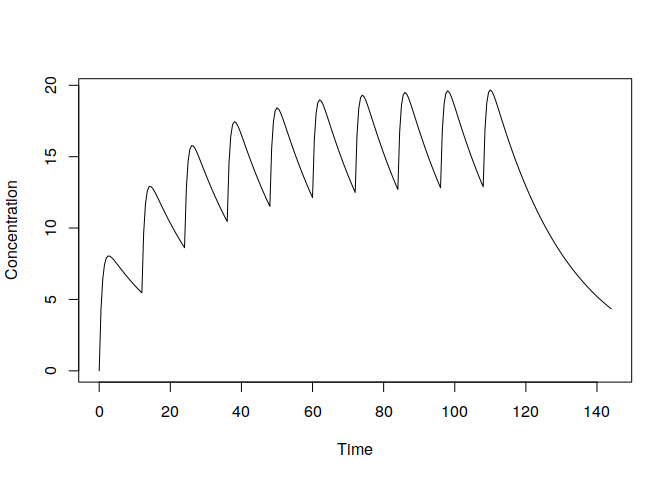
For a more detailed introduction to the package, see the vignette.
There is a companion shiny app that provides a demo of the package capabilities, and also generates code that can be placed in an R script. It can be accessed at https://benjaminrich.shinyapps.io/linpk-demo-app/ or run locally by pasting the following lines in an R console:
# Make sure the required packages are installed (if not, install them)
sapply(c("shiny", "shinyjs", "shinyAce", "dygraphs", "linpk"), requireNamespace)
linpk::linpkApp()The app will open in a browser, and looks like this:

These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.