
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.

R package for fitting and testing alternative models for single cohort litter decomposition data
#install.packages("remotes")
remotes::install_github("cornwell-lab-unsw/litterfitter")
library(litterfitter)At the moment there is one key function which is
fit_litter which can fit 6 different types of decomposition
trajectories. Note that the fitted object is a litfit
object
fit <- fit_litter(time=c(0,1,2,3,4,5,6),
mass.remaining =c(1,0.9,1.01,0.4,0.6,0.2,0.01),
model="weibull",
iters=500)
class(fit)You can visually compare the fits of different non-linear equations
with the plot_multiple_fits function:
plot_multiple_fits(time=c(0,1,2,3,4,5,6),
mass.remaining=c(1,0.9,1.01,0.4,0.6,0.2,0.01),
model=c("neg.exp","weibull"),
iters=500)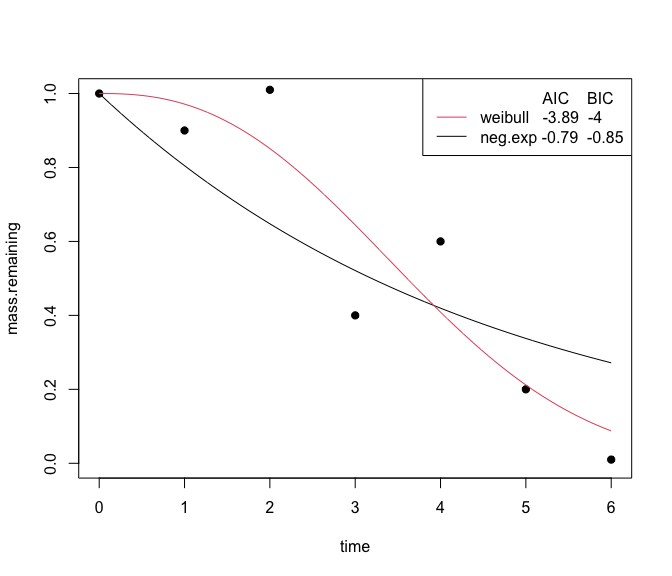
Calling plot on a litfit object will show
you the data, the curve fit, and even the equation, with the estimated
coefficients:
plot(fit)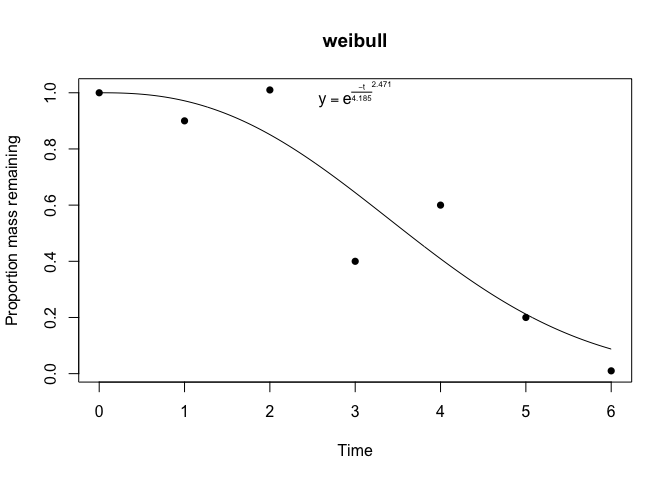
The summary of a litfit object will show you some of the
summary statistics for the fit.
#> Summary of litFit object
#> Model type: weibull
#> Number of observations: 7
#> Parameter fits: 4.19
#> Parameter fits: 2.47
#> Time to 50% mass loss: 3.61
#> Implied steady state litter mass: 3.71 in units of yearly input
#> AIC: -3.8883
#> AICc: -0.8883
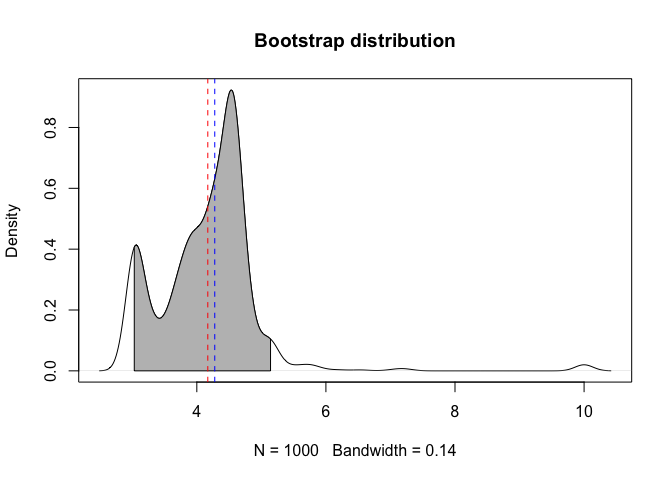
#> BIC: -3.9965From the litfit object you can then see the uncertainty
in the parameter estimate by bootstrapping
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.