
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.

Framework for building modular Monte Carlo risk analysis models. It
extends the capabilities of mc2d to facilitate working with
multiple risk pathways, variates and scenarios. It provides tools to
organize risk analysis in independent flexible modules, perform
multivariate mcnode operations, automate the creation of mcnodes and
visualize risk analysis models.
Install from CRAN:
install.packages("mcmodule")
library("mcmodule")Install latest development version from GitHub (requires
devtool package)
# install.packages("devtools")
devtools::install_github("NataliaCiria/mcmodule")example_data <- data.frame(
category_1 = c("a", "b", "a", "b"), # Category 1 (e.g., type)
category_2 = c("blue", "blue", "red", "red"), # Category 2 (e.g., group)
x_min = c(0.07, 0.3, 0.2, 0.5), # Minimum value for parameter x
x_max = c(0.08, 0.4, 0.3, 0.6), # Maximum value for parameter x
y = c(0.01, 0.02, 0.03, 0.04) # Value for parameter y
)example_data_keys <- list(
example_data = list(
cols = names(example_data), # All columns in the data
keys = c("category_1", "category_2") # Unique identifiers for each row
)
)example_mctable <- data.frame(
mcnode = c("x", "y"), # Names of the nodes
description = c("Probability x", "Probability y"), # Descriptions
mc_func = c("runif", NA), # Distribution function for x, none for y
from_variable = c(NA, NA), # Source variable (not used here)
transformation = c(NA, NA), # Transformation (not used here)
sensi_analysis = c(FALSE, FALSE) # Include in sensitivity analysis
)example_exp <- quote({
result <- x * y # Calculate result as product of x and y
})eval_module(), creating the
stochastic nodes and evaluating the expressionexample_mcmodule <- eval_module(
exp = c(example = example_exp), # Model expression(s)
data = example_data, # Input data
mctable = example_mctable, # Node definitions
data_keys = example_data_keys # Data keys for matching
)# Summarize the 'result' node
mc_summary(example_mcmodule, "result")
# Get 'result' aggregated by category 1
example_mcmodule<-example_mcmodule%>%
agg_totals(
mc_name = "result",
agg_keys = c("category_1")
)
# Print aggregated 'result'
example_mcmodule$node_list$result_agg$summary
# Visualize the mcmodule
mc_network(example_mcmodule, legend = TRUE)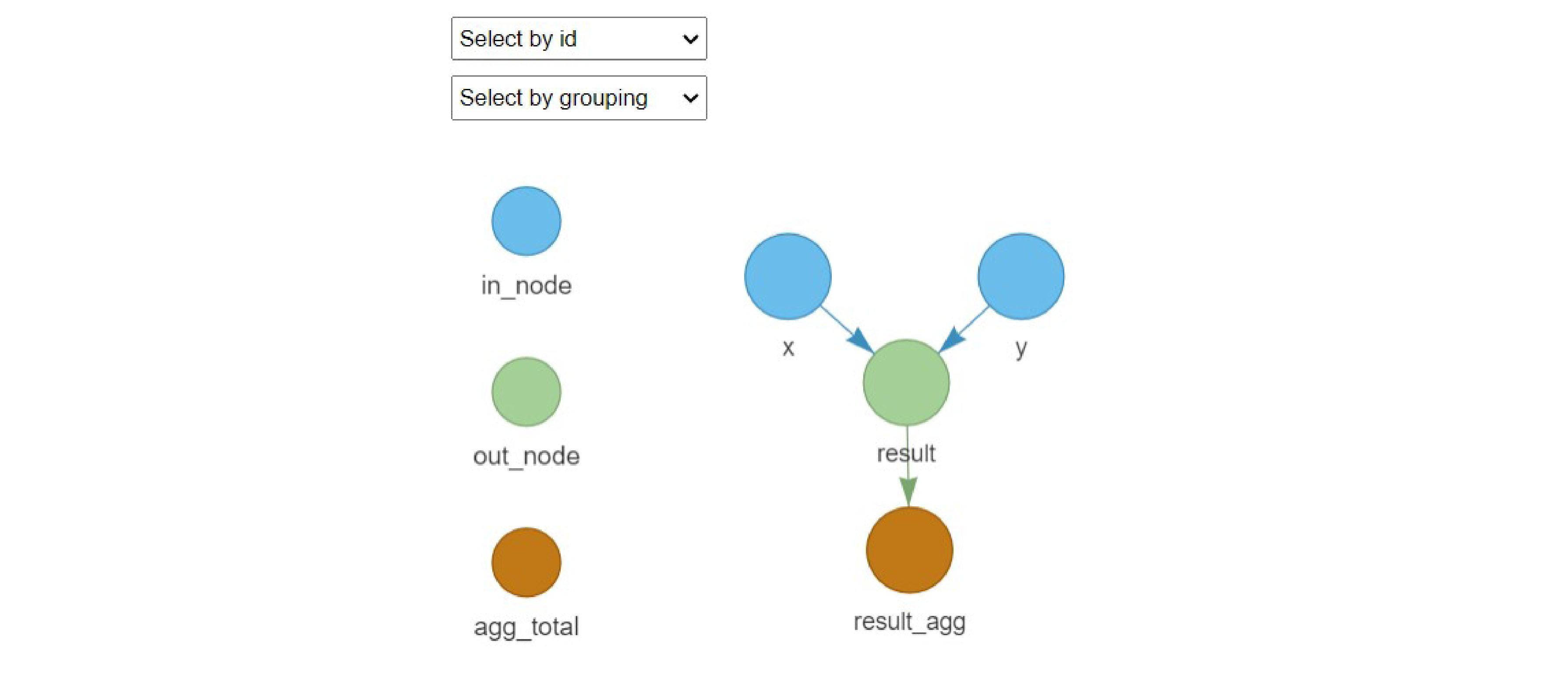
Further documentation and examples can be found in the vignette and in the introduction article.
If you use mcmodule in your research, please cite:
Ciria, N. (2025). mcmodule: Modular Monte Carlo Risk Analysis. R package version 1.1.0. https://nataliaciria.github.io/mcmodule/
mcmodule was developed with support from:
BIOSECURE, a European Union’s HORIZON Europe FARM2FORK project
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.