
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.

mcptools implements the Model Context Protocol in R. There are two sides to mcptools:
R as an MCP server:
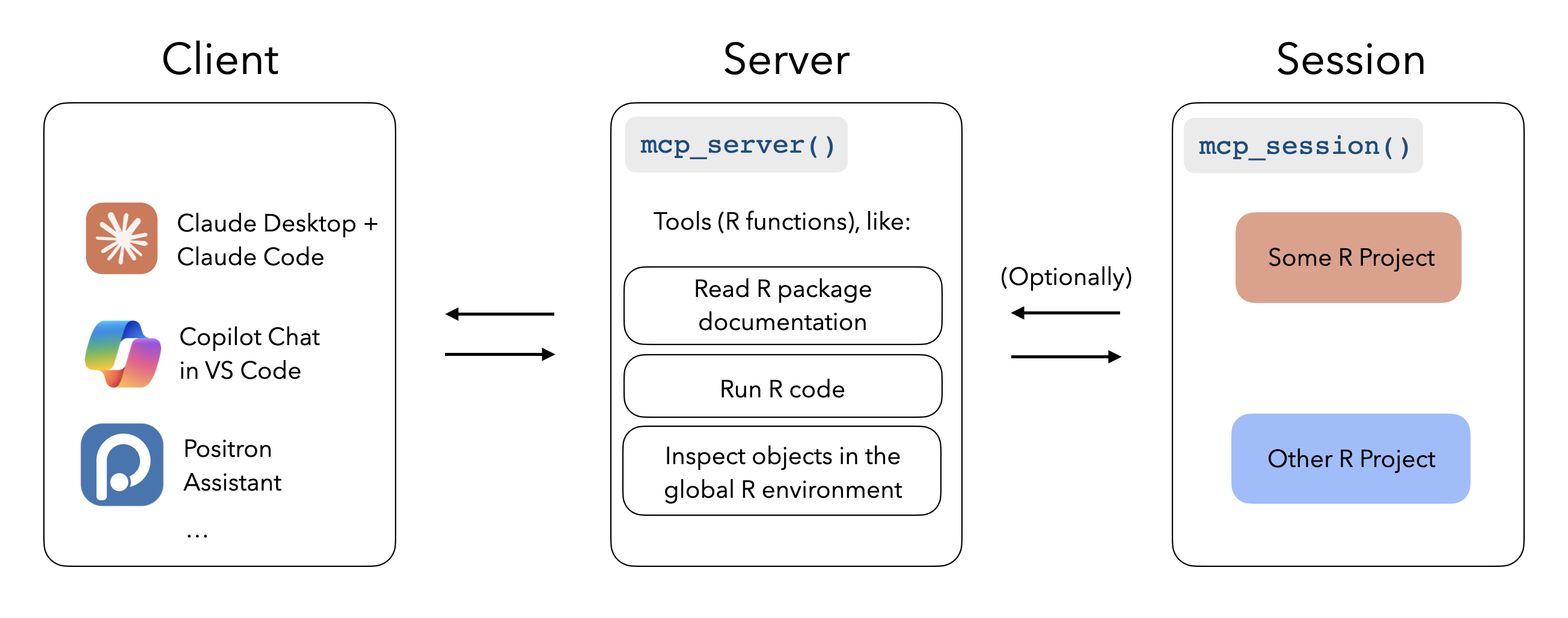
When configured with mcptools, MCP-enabled tools like Claude Desktop, Claude Code, and VS Code GitHub Copilot can run R code in the sessions you have running to answer your questions. While the package supports configuring arbitrary R functions, you may be interested in the btw package’s integrated support for mcptools, which provides a default set of tools to to peruse the documentation of packages you have installed, check out the objects in your global environment, and retrieve metadata about your session and platform.
R as an MCP client:
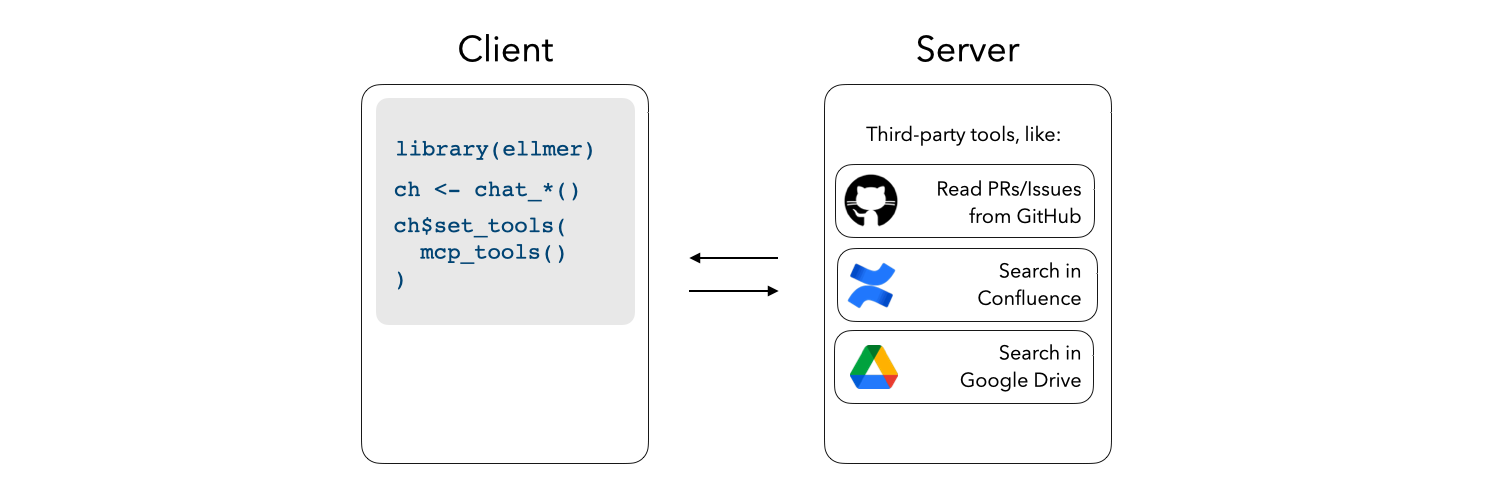
Register third-party MCP servers with ellmer chats to integrate additional context into e.g. shinychat and querychat apps.
NOTE:
This package used to be called acquaint and supplied a default set of tools from btw when R was used as an MCP server. The direction of the dependency has been reversed; to use the same functionality from before, transition
acquaint::mcp_server()tobtw::btw_mcp_server()andacquaint::mcp_session()tobtw::btw_mcp_session().
Install mcptools from CRAN with:
install.packages("mcptools")You can install the development version of mcptools like so:
pak::pak("posit-dev/mcptools")mcptools can be hooked up to any application that supports MCP. For
example, to use with Claude Desktop, you might paste the following in
your Claude Desktop configuration (on macOS, at
~/Library/Application Support/Claude/claude_desktop_config.json):
{
"mcpServers": {
"r-mcptools": {
"command": "Rscript",
"args": ["-e", "mcptools::mcp_server()"]
}
}
}Or, to use with Claude Code, you might type in a terminal:
claude mcp add -s "user" r-mcptools -- Rscript -e "mcptools::mcp_server()"Then, if you’d like models to access variables in specific R
sessions, call mcptools::mcp_session() in those sessions.
(You might include a call to this function in your .Rprofile, perhaps
using usethis::edit_r_profile(), to automatically register
every session you start up.)
mcptools uses the Claude Desktop configuration file format to register third-party MCP servers, as most MCP servers provide setup instructions for Claude Desktop in their documentation. For example, here’s what the official GitHub MCP server configuration would look like:
{
"mcpServers": {
"github": {
"command": "docker",
"args": [
"run",
"-i",
"--rm",
"-e",
"GITHUB_PERSONAL_ACCESS_TOKEN",
"ghcr.io/github/github-mcp-server"
],
"env": {
"GITHUB_PERSONAL_ACCESS_TOKEN": "<YOUR_TOKEN>"
}
}
}
}Once the configuration file has been created (by default, mcptools
will look to
file.path("~", ".config", "mcptools", "config.json")),
mcp_tools() will return a list of ellmer tools which you
can pass directly to the $set_tools() method from
ellmer:
ch <- ellmer::chat_anthropic()
ch$set_tools(mcp_tools())
ch$chat("What issues are open on posit-dev/mcptools?")In Claude Desktop, I’ll write the following:
“From what year is the earliest recorded sample in the
foresteddata in my Positron session?”
Without mcptools, Claude couldn’t get far here; by default, it can’t run R code and doesn’t have any way to “speak to” my interactive R sessions.
Using the package, the model asks to describe the data frame using a structure that will show summary statistics from the data. mcptools will appropriately route the request to the open Positron session, forwarding the results back to the model for it to situate in a response.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.