
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.

An increasingly important source of health-related bibliographic content are preprints - preliminary versions of research articles that have yet to undergo peer review. The two preprint repositories most relevant to health-related sciences are medRxiv and bioRxiv, both of which are operated by the Cold Spring Harbor Laboratory.
The goal of the medrxivr R package is two-fold. In the
first instance, it provides programmatic access to the Cold Spring Harbour Laboratory (CSHL)
API, allowing users to easily download medRxiv and bioRxiv preprint
metadata (e.g. title, abstract, publication date, author list, etc) into
R. The package also provides access to a maintained static snapshot of
the medRxiv repository (see Data sources).
Secondly, medrxivr provides functions to search the
downloaded preprint records using regular expressions and Boolean logic,
as well as helper functions that allow users to export their search
results to a .BIB file for easy import to a reference manager and to
download the full-text PDFs of preprints matching their search
criteria.
To install the stable version of the package from CRAN:
install.packages("medrxivr")
library(medrxivr)Alternatively, to install the development version from GitHub, use the following code:
install.packages("devtools")
devtools::install_github("ropensci/medrxivr")
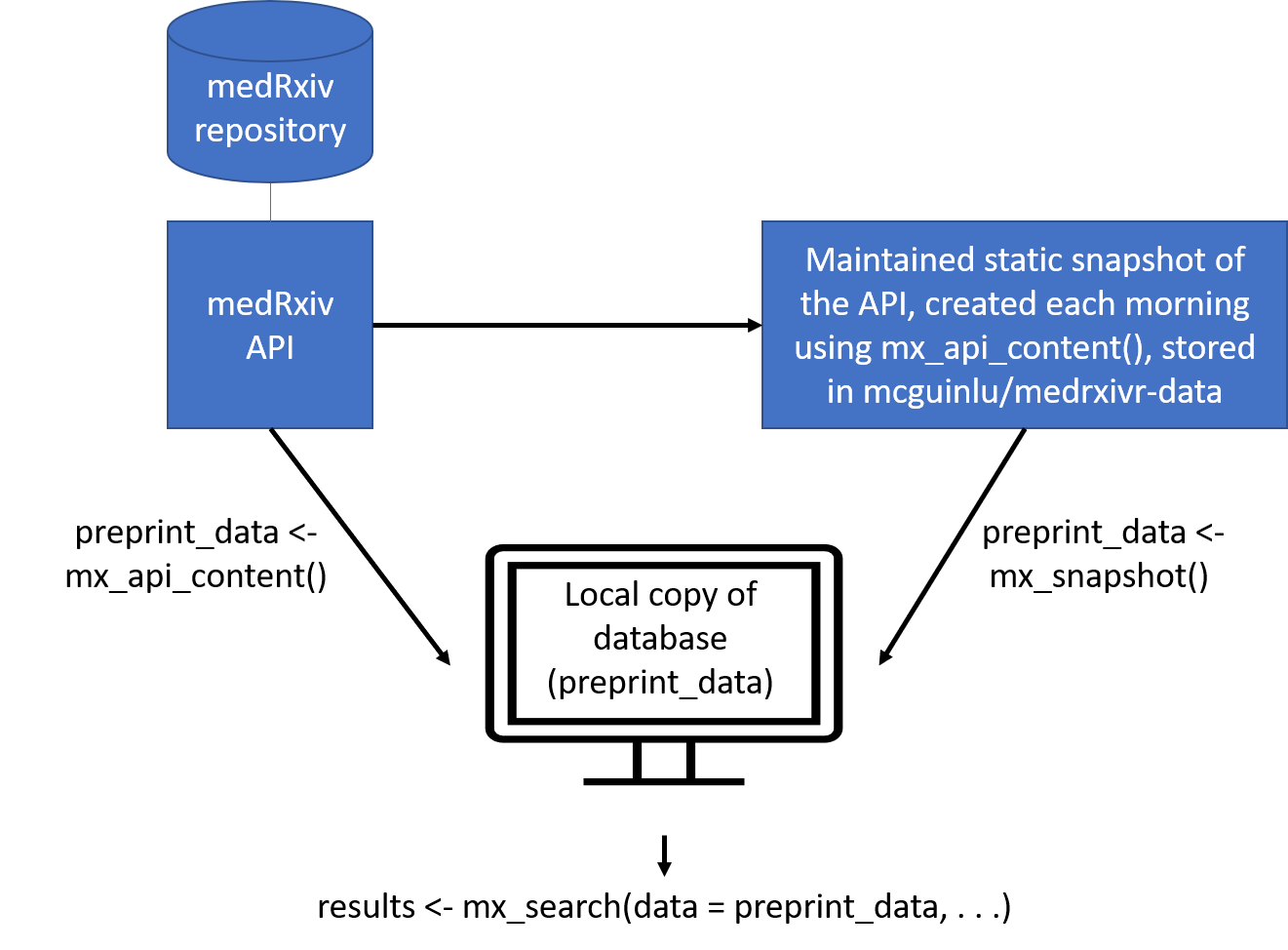
library(medrxivr)medrixvr provides two ways to access medRxiv data:
mx_api_content(server = "medrxiv") creates a local copy
of all data available from the medRxiv API at the time the function is
run.# Get a copy of the database from the live medRxiv API endpoint
preprint_data <- mx_api_content() mx_snapshot() provides access to a static snapshot of
the medRxiv database. The snapshot is created each morning at 6am using
mx_api_content() and is stored as CSV file in the medrxivr-data
repository. This method does not rely on the API (which can become
unavailable during peak usage times) and is usually faster (as it reads
data from a CSV rather than having to re-extract it from the API).
Discrepancies between the most recent static snapshot and the live
database can be assessed using mx_crosscheck().# Get a copy of the database from the daily snapshot
preprint_data <- mx_snapshot() The relationship between the two methods for the medRxiv database is summarised in the figure below:
Only one data source exists for the bioRxiv repository:
mx_api_content(server = "biorxiv") creates a local copy
of all data available from the bioRxiv API endpoint at the time the
function is run. Note: due to it’s size, downloading a
complete copy of the bioRxiv repository in this manner takes a long time
(~ 1 hour).# Get a copy of the database from the live bioRxiv API endpoint
preprint_data <- mx_api_content(server = "biorxiv")Once you have created a local copy of either the medRxiv or bioRxiv
preprint database, you can pass this object (preprint_data
in the examples above) to mx_search() to search the
preprint records using an advanced search strategy.
# Import the medrxiv database
preprint_data <- mx_snapshot()
#> Using medRxiv snapshot - 2022-07-06 01:09
# Perform a simple search
results <- mx_search(data = preprint_data,
query ="dementia")
#> Found 427 record(s) matching your search.
# Perform an advanced search
topic1 <- c("dementia","vascular","alzheimer's") # Combined with Boolean OR
topic2 <- c("lipids","statins","cholesterol") # Combined with Boolean OR
myquery <- list(topic1, topic2) # Combined with Boolean AND
results <- mx_search(data = preprint_data,
query = myquery)
#> Found 143 record(s) matching your search.You can also explore which search terms are contributing most to your
search by setting report = TRUE:
results <- mx_search(data = preprint_data,
query = myquery,
report = TRUE)
#> Found 143 record(s) matching your search.
#> Total topic 1 records: 2272
#> dementia: 427
#> vascular: 1918
#> alzheimer's: 0
#> Total topic 2 records: 410
#> lipids: 157
#> statins: 61
#> cholesterol: 255Pass the results of your search above (the results
object) to the mx_export() to export references for
preprints matching your search results to a .BIB file so that they can
be easily imported into a reference manager (e.g. Zotero, Mendeley).
mx_export(data = results,
file = "mx_search_results.bib")Pass the results of your search above (the results
object) to the mx_download() function to download a copy of
the PDF for each record found by your search.
mx_download(results, # Object returned by mx_search(), above
"pdf/", # Directory to save PDFs to
create = TRUE) # Create the directory if it doesn't existBy default, the mx_api_*() functions clean the data
returned by the API for use with other medrxivr
functions.
To access the raw data returned by the API, the clean
argument should set to FALSE:
mx_api_content(to_date = "2019-07-01", clean = FALSE)See this article for more details.
Detailed guidance, including advice on how to design complex search
strategies, is available on the medrxivr
website.
See here for the code used to take the
daily snapshot and the code that powers the
medrxivr web app.
The focus of medrxivr is on providing tools to allow
users to import and then search medRxiv and bioRxiv data. Below are a
list of complementary packages that provide distinct but related
functionality when working with medRxiv and bioRxiv data:
rbiorxiv
by Nicholas Fraser
provides access to the same medRxiv and bioRxiv content data as
medrxivr, but also provides access to the usage
data (e.g. downloads per month) that the Cold Spring Harbour Laboratory
API offers. This is useful if you wish to explore, for example, how
the number of PDF downloads from bioRxiv has grown over time.Please note that this package is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
This package and the data it accesses/returns are provided “as is”, with no guarantee of accuracy. Please be sure to check the accuracy of the data yourself (and do let me know if you find an issue so I can fix it for everyone!)
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.