The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
The goal of mixdir is to cluster high dimensional categorical datasets.
It can
mixdir(select_latent=TRUE))A detailed description of the algorithm and the features of the package can be found in the the accompanying paper. If you find the package useful please cite
C. Ahlmann-Eltze and C. Yau, “MixDir: Scalable Bayesian Clustering for High-Dimensional Categorical Data”, 2018 IEEE 5th International Conference on Data Science and Advanced Analytics (DSAA), Turin, Italy, 2018, pp. 526-539.
install.packages("mixdir")
# Or to get the latest version from github
devtools::install_github("const-ae/mixdir")Clustering the mushroom data set.

# Loading the library and the data
library(mixdir)
set.seed(1)
data("mushroom")
# High dimensional dataset: 8124 mushroom and 23 different features
mushroom[1:10, 1:5]
#> bruises cap-color cap-shape cap-surface edible
#> 1 bruises brown convex smooth poisonous
#> 2 bruises yellow convex smooth edible
#> 3 bruises white bell smooth edible
#> 4 bruises white convex scaly poisonous
#> 5 no gray convex smooth edible
#> 6 bruises yellow convex scaly edible
#> 7 bruises white bell smooth edible
#> 8 bruises white bell scaly edible
#> 9 bruises white convex scaly poisonous
#> 10 bruises yellow bell smooth edibleCalling the clustering function mixdir on a subset of
the data:
# Clustering into 3 latent classes
result <- mixdir(mushroom[1:1000, 1:5], n_latent=3)Analyzing the result
# Latent class of of first 10 mushrooms
head(result$pred_class, n=10)
#> [1] 3 1 1 3 2 1 1 1 3 1
# Soft Clustering for first 10 mushrooms
head(result$class_prob, n=10)
#> [,1] [,2] [,3]
#> [1,] 3.103495e-07 1.055098e-05 9.999891e-01
#> [2,] 9.998594e-01 4.683764e-06 1.359291e-04
#> [3,] 9.998944e-01 3.111462e-06 1.025194e-04
#> [4,] 5.778033e-04 7.114603e-08 9.994221e-01
#> [5,] 3.662625e-07 9.999992e-01 4.183025e-07
#> [6,] 9.996461e-01 8.764031e-08 3.537838e-04
#> [7,] 9.998944e-01 3.111462e-06 1.025194e-04
#> [8,] 9.997331e-01 5.822320e-08 2.668420e-04
#> [9,] 5.778033e-04 7.114603e-08 9.994221e-01
#> [10,] 9.999999e-01 5.850067e-09 9.845112e-08
pheatmap::pheatmap(result$class_prob, cluster_cols=FALSE,
labels_col = paste("Class", 1:3))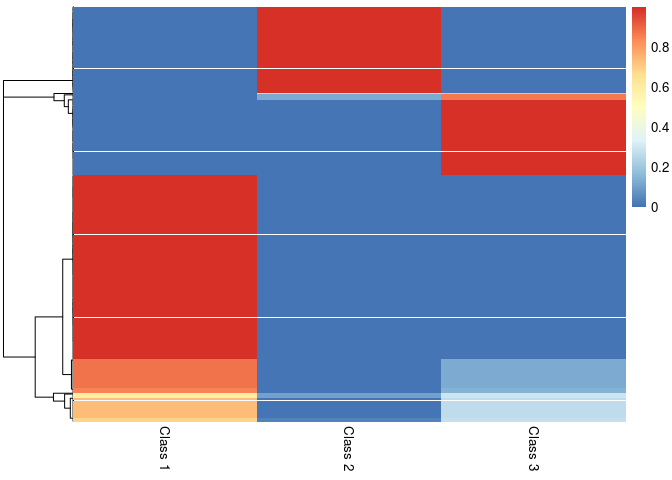
# Structure of latent class 1
# (bruises, cap color either yellow or white, edible etc.)
purrr::map(result$category_prob, 1)
#> $bruises
#> bruises no
#> 0.9998223256 0.0001776744
#>
#> $`cap-color`
#> brown gray red white yellow
#> 0.0001775934 0.0001819672 0.0001776373 0.4079822666 0.5914805356
#>
#> $`cap-shape`
#> bell convex flat sunken
#> 0.3926736 0.4767291 0.1304197 0.0001776
#>
#> $`cap-surface`
#> fibrous scaly smooth
#> 0.0568571 0.4871396 0.4560033
#>
#> $edible
#> edible poisonous
#> 0.9998223174 0.0001776826
# The most predicitive features for each class
find_predictive_features(result, top_n=3)
#> column answer class probability
#> 19 cap-color yellow 1 0.9993990
#> 22 cap-shape bell 1 0.9990947
#> 1 bruises bruises 1 0.7089533
#> 48 edible poisonous 3 0.9980468
#> 15 cap-color red 3 0.8462032
#> 9 cap-color brown 3 0.6473043
#> 5 bruises no 2 0.9990364
#> 11 cap-color gray 2 0.9978218
#> 32 cap-shape sunken 2 0.9936162
# For example: if all I know about a mushroom is that it has a
# yellow cap, then I am 99% certain that it will be in class 1
predict(result, c(`cap-color`="yellow"))
#> [,1] [,2] [,3]
#> [1,] 0.999399 0.0003004692 0.0003004907
# Note the most predictive features are different from the most typical ones
find_typical_features(result, top_n=3)
#> column answer class probability
#> 1 bruises bruises 1 0.9998223
#> 43 edible edible 1 0.9998223
#> 19 cap-color yellow 1 0.5914805
#> 3 bruises bruises 3 0.9995546
#> 27 cap-shape convex 3 0.7460615
#> 9 cap-color brown 3 0.6746224
#> 44 edible edible 2 0.9995310
#> 5 bruises no 2 0.9713177
#> 35 cap-surface fibrous 2 0.7355413Dimensionality Reduction
# Defining Features
def_feat <- find_defining_features(result, mushroom[1:1000, 1:5], n_features = 3)
print(def_feat)
#> $features
#> [1] "cap-color" "bruises" "edible"
#>
#> $quality
#> [1] 74.35146
# Plotting the most important features gives an immediate impression
# how the cluster differ
plot_features(def_feat$features, result$category_prob)
#> Loading required namespace: ggplot2
#> Loading required namespace: tidyr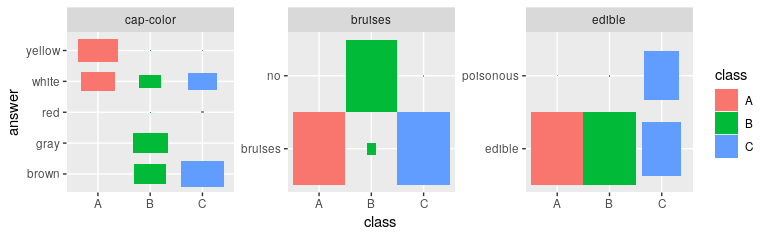
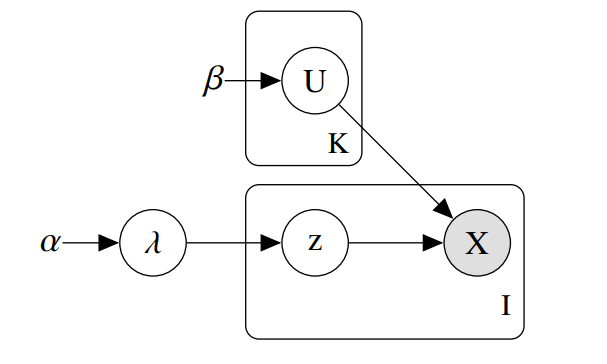
The package implements a variational inference algorithm to solve a Bayesian latent class model (LCM).

These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.