
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.

The {optimizeR} package provides an object-oriented
framework for optimizer functions in R and offers some convenience when
minimizing or maximizing.
❌ You won’t need this package if you…
{optimx},
a framework to compare about 30 optimizers,✅ But you might find the package useful if you want to…
{optimx} or other frameworks; see the CRAN Task View:
Optimization and Mathematical Programming for an overview),The following demo is a bit artificial but showcases the package purpose. Let’s assume we want to
stats::nlm and pracma::nelder_mead.We can do this task with {optimizeR}. You can install
the released package version from CRAN with:
install.packages("optimizeR")Then load the package via library("optimizeR") and you
should be ready to go.
1. Define the objective function
Let \(f:\mathbb{R}^4\to\mathbb{R}\) with
f <- function(a, b, x, y) {
a * exp(-0.2 * sqrt(0.5 * (x^2 + y^2))) + exp(0.5 * (cos(2 * pi * x) + cos(2 * pi * y))) - exp(1) - b
}For a = b = 20, this is the negative Ackley function
with a global maximum in x = y = 0:
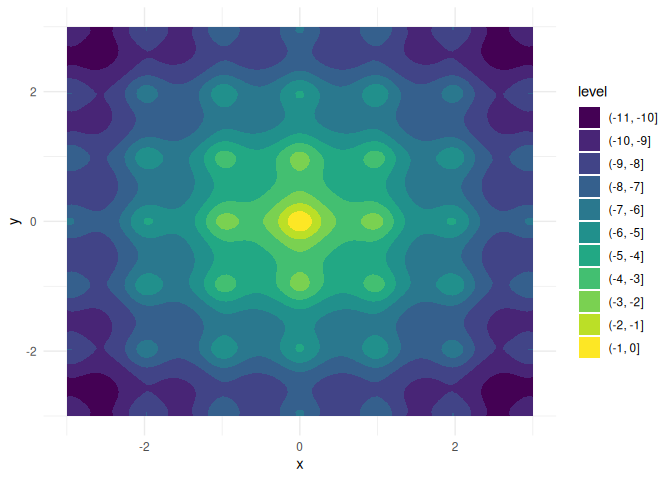
We want to keep a and b fixed here and
optimize over x and y (which are also both
single numeric values).
Two problems would occur if we would optimize f with say
stats::nlm directly:
x and y)
andBoth artifacts are not allowed by stats::nlm and most of
other available optimizers, but supported by {optimizeR}.
We just have to define an objective object which we later can pass to
the optimizers:
objective <- Objective$new(
f = f, # f is our objective function
target = c("x", "y"), # x and y are the target arguments
npar = c(1, 1), # the target arguments have both a length of 1
"a" = 20,
"b" = 20 # a and b have fixed values
)2. Create the optimizer objects
Now that we have defined the objective function, let’s define our
optimizer objects. For stats::nlm, this is a one-liner:
nlm <- Optimizer$new(which = "stats::nlm")The {optimizeR} package provides a dictionary of
optimizers, that can be directly selected via the which
argument. For an overview of available optimizers, see:
optimizer_dictionary
#> <Dictionary> optimization algorithms
#> Keys:
#> - lbfgsb3c::lbfgsb3c
#> - lbfgsb3c::lbfgsb3
#> - lbfgsb3c::lbfgsb3f
#> - lbfgsb3c::lbfgsb3x
#> - stats::nlm
#> - stats::nlminb
#> - stats::optim
#> - ucminf::ucminfBut in fact, any optimizer that is not contained in the dictionary
can be put into the {optimizeR} framework by setting
which = "custom" first…
nelder_mead <- Optimizer$new(which = "custom")
#> Use method `$definition()` next to define a custom optimizer.… and using the $definition() method next:
nelder_mead$definition(
algorithm = pracma::nelder_mead, # the optimization function
arg_objective = "fn", # the argument name for the objective function
arg_initial = "x0", # the argument name for the initial values
out_value = "fmin", # the element for the optimal function value in the output
out_parameter = "xmin", # the element for the optimal parameters in the output
direction = "min" # the optimizer minimizes
)3. Set a time limit
Each optimizer object has a field called $seconds which
equals Inf by default. You can optionally set a different,
single numeric value here to set a time limit in seconds for the
optimization:
nlm$seconds <- 10
nelder_mead$seconds <- 10Note that not everything (especially compiled C code) can technically
be timed out, see the help site
help("withTimeout", package = "R.utils") for more
details.
4. Maximize the objective function
Each optimizer object has the two methods $maximize()
and $minimize() for function maximization or minimization,
respectively. Both methods require values for the two arguments
objective (either an objective object as defined above
or just a function) andinitial (an initial parameter vector from where the
optimizer should start)and optionally accepts additional arguments to be passed to the optimizer or the objective function.
nlm$maximize(objective = objective, initial = c(3, 3))
#> $value
#> [1] -6.559645
#>
#> $parameter
#> [1] 1.974451 1.974451
#>
#> $seconds
#> [1] 0.0242281
#>
#> $initial
#> [1] 3 3
#>
#> $error
#> [1] FALSE
#>
#> $gradient
#> [1] 5.757896e-08 5.757896e-08
#>
#> $code
#> [1] 1
#>
#> $iterations
#> [1] 6nelder_mead$maximize(objective = objective, initial = c(3, 3))
#> $value
#> [1] 0
#>
#> $parameter
#> [1] 0 0
#>
#> $seconds
#> [1] 0.008966923
#>
#> $initial
#> [1] 3 3
#>
#> $error
#> [1] FALSE
#>
#> $count
#> [1] 105
#>
#> $info
#> $info$solver
#> [1] "Nelder-Mead"
#>
#> $info$restarts
#> [1] 0Note that
the inputs for the objective function and initial parameter values are named consistently across optimizers,
the output values for the optimal parameter vector and the maximimum function value are also named consistently across optimizers,
the output contains the initial parameter values and the optimization time in seconds and additionally other optimizer-specific elements,
pracma::nelder_mead outperforms
stats::nlm here both in terms of optimization time and
convergence to the global maximum.
If you have any questions, found a bug, need a feature, just file an issue on GitHub.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.