
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.

This package offers convenient access to a new and extensive database containing a list of 7892 endemic plant species found in Peru. This comprehensive collection provides detailed botanical information, including accepted names, family, genus, species, infraspecific taxonomy, authorship, publication details, and temporal information including both actual and nominal publication years for each species.
The construction of the ppendemic package is built upon
valuable data sourced from the renowned World Checklist of
Vascular Plants (WCVP) database. The WCVP is an international
collaborative programme initiated in 1988 by Rafaël Govaerts that
provides high-quality expert-reviewed taxonomic data on all vascular
plants. As a highly authoritative resource updated daily, WCVP offers
comprehensive information on plant taxonomy and occurrence worldwide,
serving as the taxonomic backbone for World Flora Online (WFO) and being
incorporated into the Catalogue of Life Checklist via GBIF. Leveraging
this data, the ppendemic package aims to present an up-to-date and novel
compilation of Peru’s endemic plant species, tailored to the diverse
ecosystems of the region.
By incorporating meticulously curated data from WCVP following the International Code of Nomenclature for algae, fungi, and plants (ICN), this package offers users a reliable and accurate resource to explore, analyze, and gain deeper insights into the rich diversity of Peru’s endemic flora. The latest version (V-15, dated 06-01-2026) includes enhanced temporal bibliographic information with sophisticated year extraction capabilities, distinguishing between actual and nominal publication years for improved citation accuracy.
Representing a significant advancement in our understanding of Peru’s endemic plant species, the ppendemic package update the previously known list of 5,507 species presented in the Red Book of Endemic Plants of Peru, bringing the total to an impressive 7892 species. This substantial increase in documented endemic species is a testament to the continuous integration of updated taxonomic data and the commitment to presenting the most current information available. With this expanded and current database, researchers, conservationists, and nature enthusiasts alike can now delve into a more comprehensive and accurate account of Peru’s unique and diverse plant biodiversity.
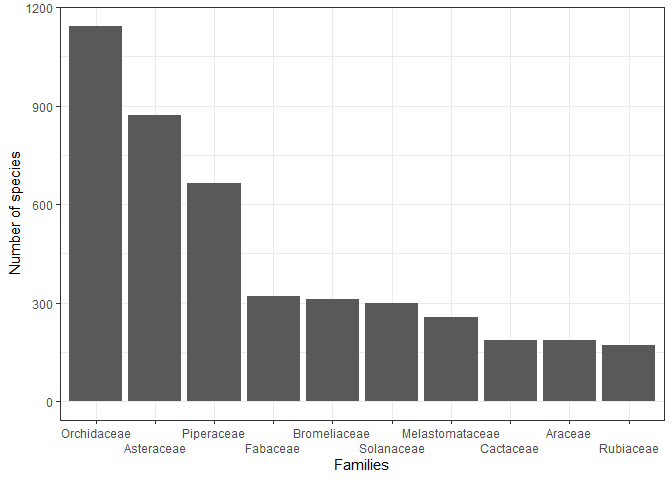
The database spans a total of 165 families, with particular richness observed in the Orchidaceae, Asteraceae, Piperaceae, Fabaceae, Bromeliaceae, Solanaceae, Melastomataceae, Cactaceae, Araceae, Rubiaceae families, all of which boast the highest number of endemic species in Peru. The enhanced dataset now includes 159 records where actual and nominal publication years differ, providing valuable insights into historical botanical publishing practices.
You can install the ppendemic package from CRAN
using:
install.packages("ppendemic")
# or
pak::pak("ppendemic")Also you can install the ppendemic package from GitHub
using the following command:
pak::pak("PaulESantos/ppendemic")After installing the ppendemic package, you can load it
into your R session using:
library(ppendemic)
#> ── Access Peruvian plant endemic data ─────────────────────── ppendemic 0.2.0 ──is_ppendemic() to check if taxa are endemicsplist <- c("Aa aurantiaca",
"Aa aurantiaaia",
"Werneria nubigena",
"Dasyphyllum brasiliense var. barnadesioides",
"Miconia firma",
"Festuca densiflora")
is_ppendemic(splist)
#> [1] "Endemic" "Endemic" "Not endemic" "Endemic" "Endemic"
#> [6] "Endemic"is_ppendemic() function is designed to work
seamlessly with tibbles, allowing users to easily analyze and determine
the endemic status of species within a tabular format.
tibble::tibble(splist = splist) |>
dplyr::mutate(endemic = is_ppendemic(splist))
#> # A tibble: 6 × 2
#> splist endemic
#> <chr> <chr>
#> 1 Aa aurantiaca Endemic
#> 2 Aa aurantiaaia Endemic
#> 3 Werneria nubigena Not endemic
#> 4 Dasyphyllum brasiliense var. barnadesioides Endemic
#> 5 Miconia firma Endemic
#> 6 Festuca densiflora EndemicTo cite the ppendemic package, please use:
citation("ppendemic")
#> To cite ppendemic in publications use:
#>
#> Santos-Andrade PE, Vilca-Bustamante LL (2025). ppendemic: A glimpse
#> at the diversity of Peru's endemic plants.
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Manual{,
#> author = {Paul E. Santos Andrade and Lucely L. Vilca Bustamante},
#> title = {ppendemic: A glimpse at the diversity of Peru's endemic plants},
#> year = {2025},
#> doi = {10.5281/zenodo.5106619},
#> url = {https://paulesantos.github.io/ppendemic/},
#> }These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.