The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
proxyC computes proximity between rows or columns of large matrices efficiently in C++. It is optimized for large sparse matrices using the Armadillo and Intel TBB libraries. Among several built-in similarity/distance measures, computation of correlation, cosine similarity and Euclidean distance is particularly fast.
This code was originally written for quanteda to compute similarity/distance between documents or features in large corpora, but separated as a stand-alone package to make it available for broader data scientific purposes.
Since proxyC v0.4.0, it requires the Intel oneAPI Threading Building Blocks for parallel computing. Windows and Mac users can download a binary package from CRAN, but Linux users must install the library by executing the commands below:
# Fedora, CentOS, RHEL
sudo yum install tbb-devel
# Debian and Ubuntu
sudo apt install libtbb-devinstall.packages("proxyC")require(Matrix)
## Loading required package: Matrix
require(microbenchmark)
## Loading required package: microbenchmark
require(ggplot2)
## Loading required package: ggplot2
require(magrittr)
## Loading required package: magrittr
# Set number of threads
options("proxyC.threads" = 8)
# Make a matrix with 99% zeros
sm1k <- rsparsematrix(1000, 1000, 0.01) # 1,000 columns
sm10k <- rsparsematrix(1000, 10000, 0.01) # 10,000 columns
# Convert to dense format
dm1k <- as.matrix(sm1k)
dm10k <- as.matrix(sm10k)With sparse matrices, proxyC is roughly 10 to 100 times faster than proxy.
bm1 <- microbenchmark(
"proxy 1k" = proxy::simil(dm1k, method = "cosine"),
"proxyC 1k" = proxyC::simil(sm1k, margin = 2, method = "cosine"),
"proxy 10k" = proxy::simil(dm10k, method = "cosine"),
"proxyC 10k" = proxyC::simil(sm10k, margin = 2, method = "cosine"),
times = 10
)
autoplot(bm1)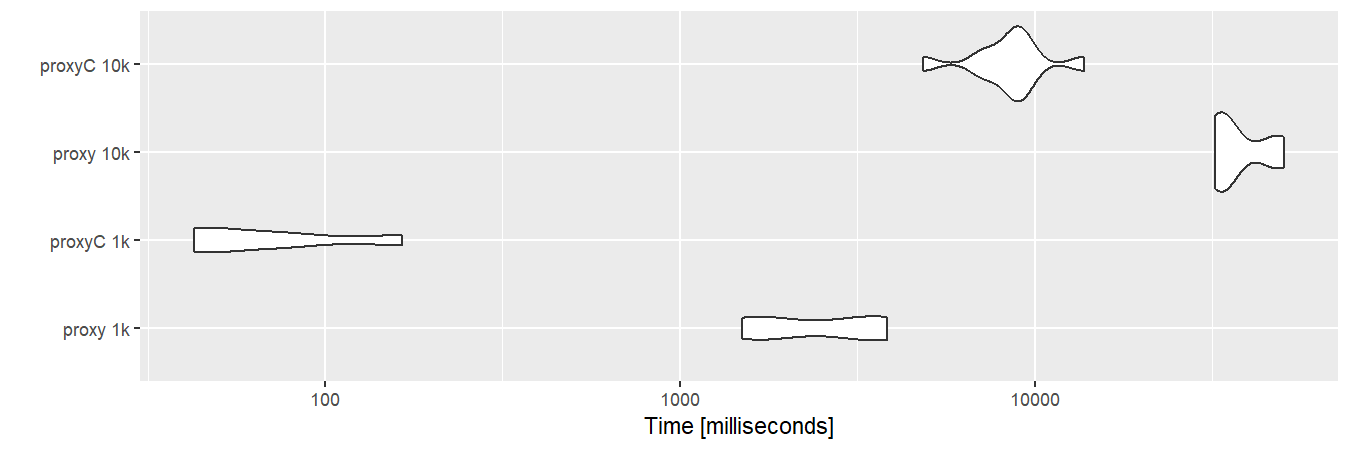
If min_simil is used, proxyC becomes
even faster because small similarity scores are floored to zero.
bm2 <- microbenchmark(
"proxyC all" = proxyC::simil(sm1k, margin = 2, method = "cosine"),
"proxyC min_simil" = proxyC::simil(sm1k, margin = 2, method = "cosine", min_simil = 0.9),
times = 10
)
autoplot(bm2)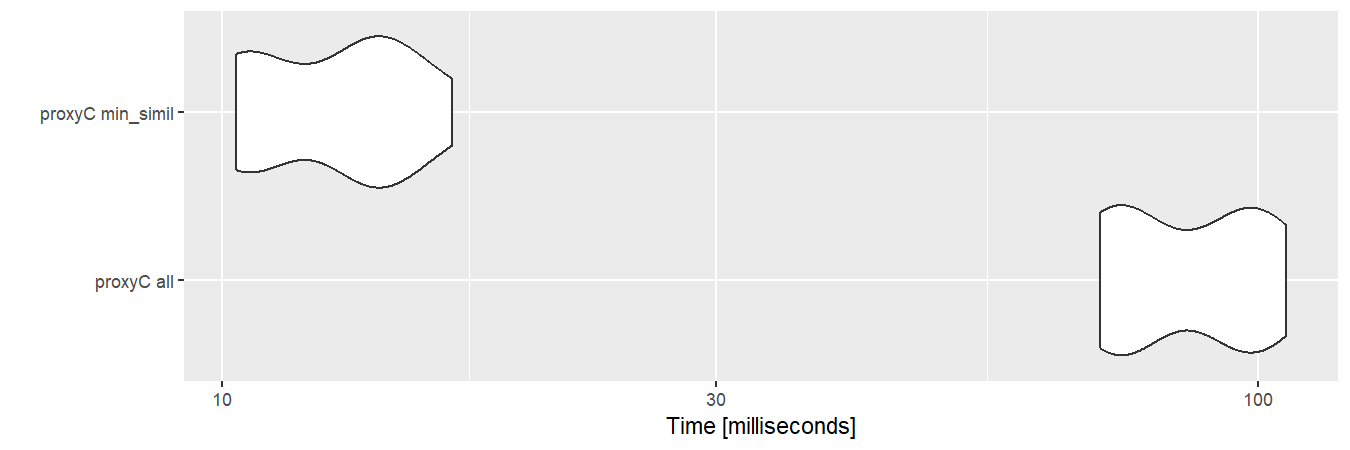
Flooring by min_simil makes the resulting object much
smaller.
proxyC::simil(sm10k, margin = 2, method = "cosine") %>%
object.size() %>%
print(units = "MB")
## 763 Mb
proxyC::simil(sm10k, margin = 2, method = "cosine", min_simil = 0.9) %>%
object.size() %>%
print(units = "MB")
## 0.2 MbIf rank is used, proxyC only returns
top-n values.
bm3 <- microbenchmark(
"proxyC rank" = proxyC::simil(sm1k, margin = 2, method = "correlation", rank = 10),
"proxyC all" = proxyC::simil(sm1k, margin = 2, method = "correlation"),
times = 10
)
autoplot(bm3)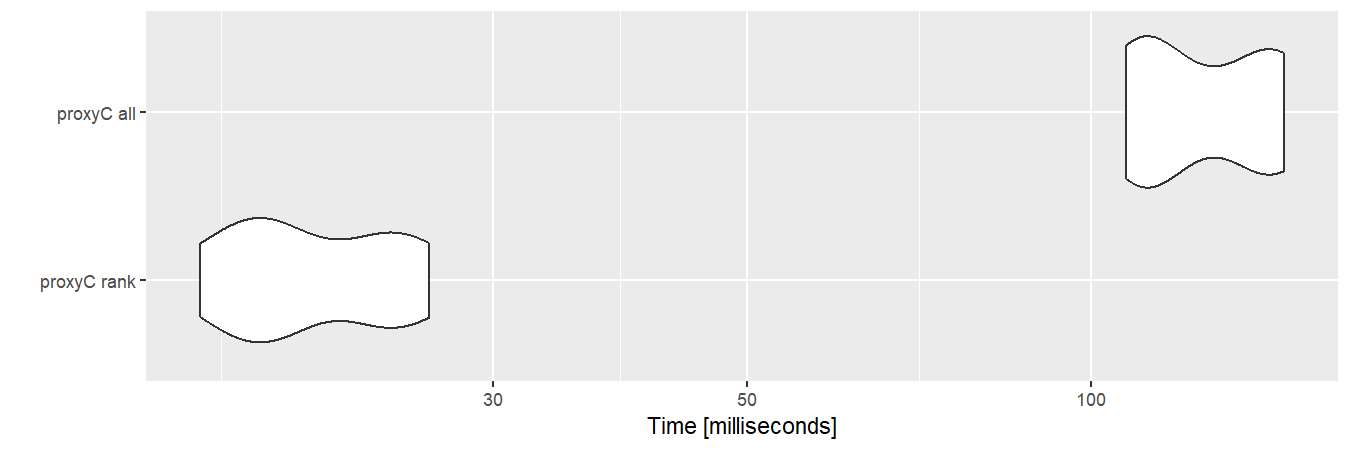
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.