The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
The goal of psc is to compare a dataset of observations against a parametric model
You can install the development version of psc from GitHub with:
# install.packages("devtools")
devtools::install_github("richJJackson/psc")This is a basic example which shows you how to solve a common problem:
library(psc)
#> Loading required package: survival
#> Loading required package: ggplot2
library(survival)
## basic example code
### Load model
data("surv.mod")
### Load Data
data("data")
#> Warning in data("data"): data set 'data' not found
### Use 'pscfit' to compare
surv.psc <- pscfit(surv.mod,data)
#> Warning in pscData_match(CFM$cov_class, CFM$cov_lev, DCcov): vi specified as a character in the model, consider respecifying
#> as a factor to ensure categories match between CFM and DC
#> Warning in pscData_match(CFM$cov_class, CFM$cov_lev, DCcov): allmets specified as a character in the model, consider respecifying
#> as a factor to ensure categories match between CFM and DCYou can use standard commands for getting a summary of your analysis…
summary(surv.psc)
#> Counterfactual Model (CFM):
#> A model of class 'flexsurvreg'
#> Fit with 3 internal knots
#>
#> CFM Formula:
#> Surv(time, cen) ~ vi/age60 + ecog + allmets + logafp + alb +
#> logcreat + logast + aet
#> <environment: 0x116b6fd60>
#>
#> CFM Summary:
#> Expected response for the outcome under the CFM:
#> S lo hi
#> 9.694 9.094 11.013
#>
#> Observed outcome from the Data Cohort:
#> [,1]
#> median 6.366
#> 0.95LCL 5.436
#> 0.95UCL 9.094
#>
#> MCMC Fit:
#> Posterior Distribution obtaine with fit summary:
#> variable rhat ess_bulk ess_tail mcse_mean
#> [1,] beta_1 1.001575 1157.117 1022.026 0.002900963
#>
#> Summary:
#> Posterior Distribution for beta:Call:
#> CFM model + beta
#>
#> Coefficients:
#> variable mean sd median q5
#> posterior beta_1 0.3707433 0.09903147 0.3708735 0.2103505
#> q95
#> posterior 0.5420673… and to see a plot of what you have done
#> variable mean sd median q5
#> posterior beta_1 0.3707433 0.09903147 0.3708735 0.2103505
#> q95
#> posterior 0.5420673
#> Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` instead.
#> ℹ The deprecated feature was likely used in the ggpubr package.
#> Please report the issue at <https://github.com/kassambara/ggpubr/issues>.
#> This warning is displayed once every 8 hours.
#> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
#> generated.
#> Ignoring unknown labels:
#> • colour : "Strata"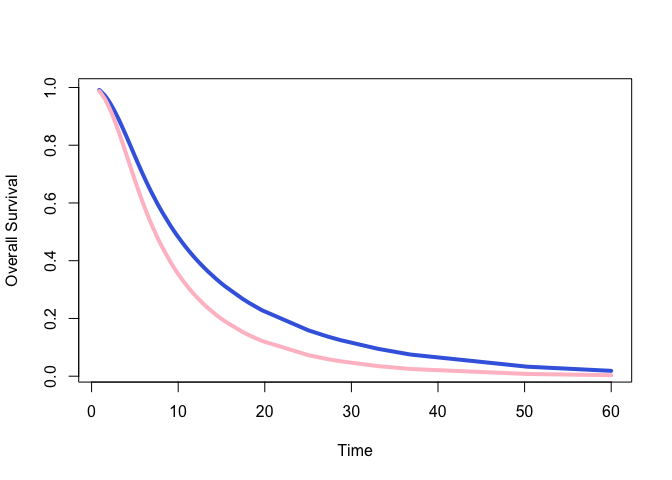
In that case, don’t forget to commit and push the resulting figure files, so they display on GitHub and CRAN.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.