The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
Quick serialization of R objects
qs provides an interface for quickly saving and reading
objects to and from disk. The goal of this package is to provide a
lightning-fast and complete replacement for the saveRDS and
readRDS functions in R.
Inspired by the fst package, qs uses a
similar block-compression design using either the lz4 or
zstd compression libraries. It differs in that it applies a
more general approach for attributes and object references.
saveRDS and readRDS are the standard for
serialization of R data, but these functions are not optimized for
speed. On the other hand, fst is extremely fast, but only
works on data.frame’s and certain column types.
qs is both extremely fast and general: it can serialize
any R object like saveRDS and is just as fast and sometimes
faster than fst.
library(qs)
df1 <- data.frame(x = rnorm(5e6), y = sample(5e6), z=sample(letters, 5e6, replace = T))
qsave(df1, "myfile.qs")
df2 <- qread("myfile.qs")# CRAN version
install.packages("qs")
# CRAN version compile from source (recommended)
remotes::install_cran("qs", type = "source", configure.args = "--with-simd=AVX2")The table below compares the features of different serialization approaches in R.
| qs | fst | saveRDS | |
|---|---|---|---|
| Not Slow | ✔ | ✔ | ❌ |
| Numeric Vectors | ✔ | ✔ | ✔ |
| Integer Vectors | ✔ | ✔ | ✔ |
| Logical Vectors | ✔ | ✔ | ✔ |
| Character Vectors | ✔ | ✔ | ✔ |
| Character Encoding | ✔ | (vector-wide only) | ✔ |
| Complex Vectors | ✔ | ❌ | ✔ |
| Data.Frames | ✔ | ✔ | ✔ |
| On disk row access | ❌ | ✔ | ❌ |
| Random column access | ❌ | ✔ | ❌ |
| Attributes | ✔ | Some | ✔ |
| Lists / Nested Lists | ✔ | ❌ | ✔ |
| Multi-threaded | ✔ | ✔ | ❌ |
qs also includes a number of advanced features:
qs implements byte shuffling filters (adopted from the
Blosc meta-compression library). These filters utilize extended CPU
instruction sets (either SSE2 or AVX2).qs also efficiently serializes S4 objects,
environments, and other complex objects.These features have the possibility of additionally increasing performance by orders of magnitude, for certain types of data. See sections below for more details.
The following benchmarks were performed comparing qs,
fst and saveRDS/readRDS in base R
for serializing and de-serializing a medium sized
data.frame with 5 million rows (approximately 115 Mb in
memory):
data.frame(a = rnorm(5e6),
b = rpois(5e6, 100),
c = sample(starnames$IAU, 5e6, T),
d = sample(state.name, 5e6, T),
stringsAsFactors = F)qs is highly parameterized and can be tuned by the user
to extract as much speed and compression as possible, if desired. For
simplicity, qs comes with 4 presets, which trades speed and
compression ratio: “fast”, “balanced”, “high” and “archive”.
The plots below summarize the performance of saveRDS,
qs and fst with various parameters:
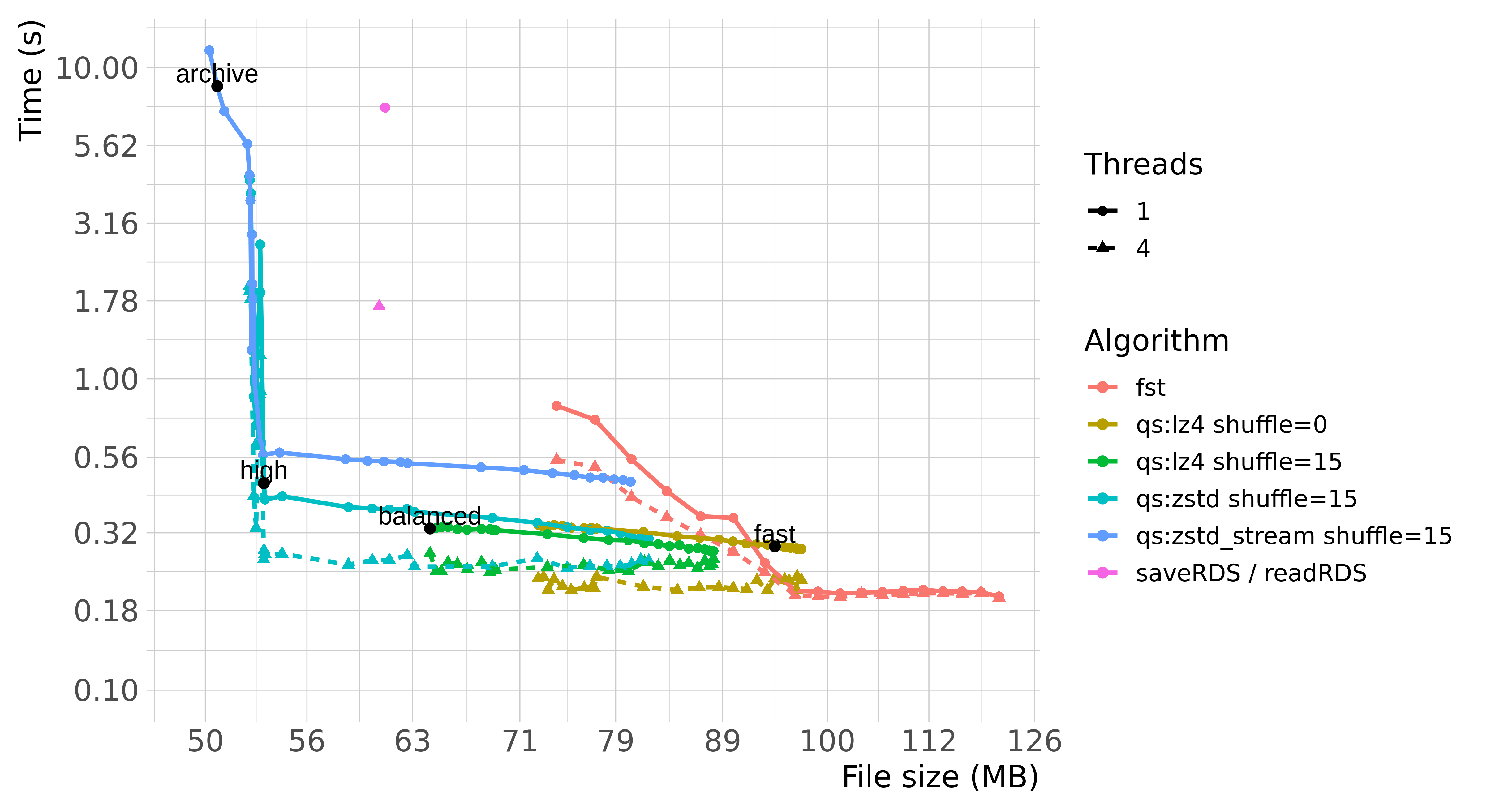
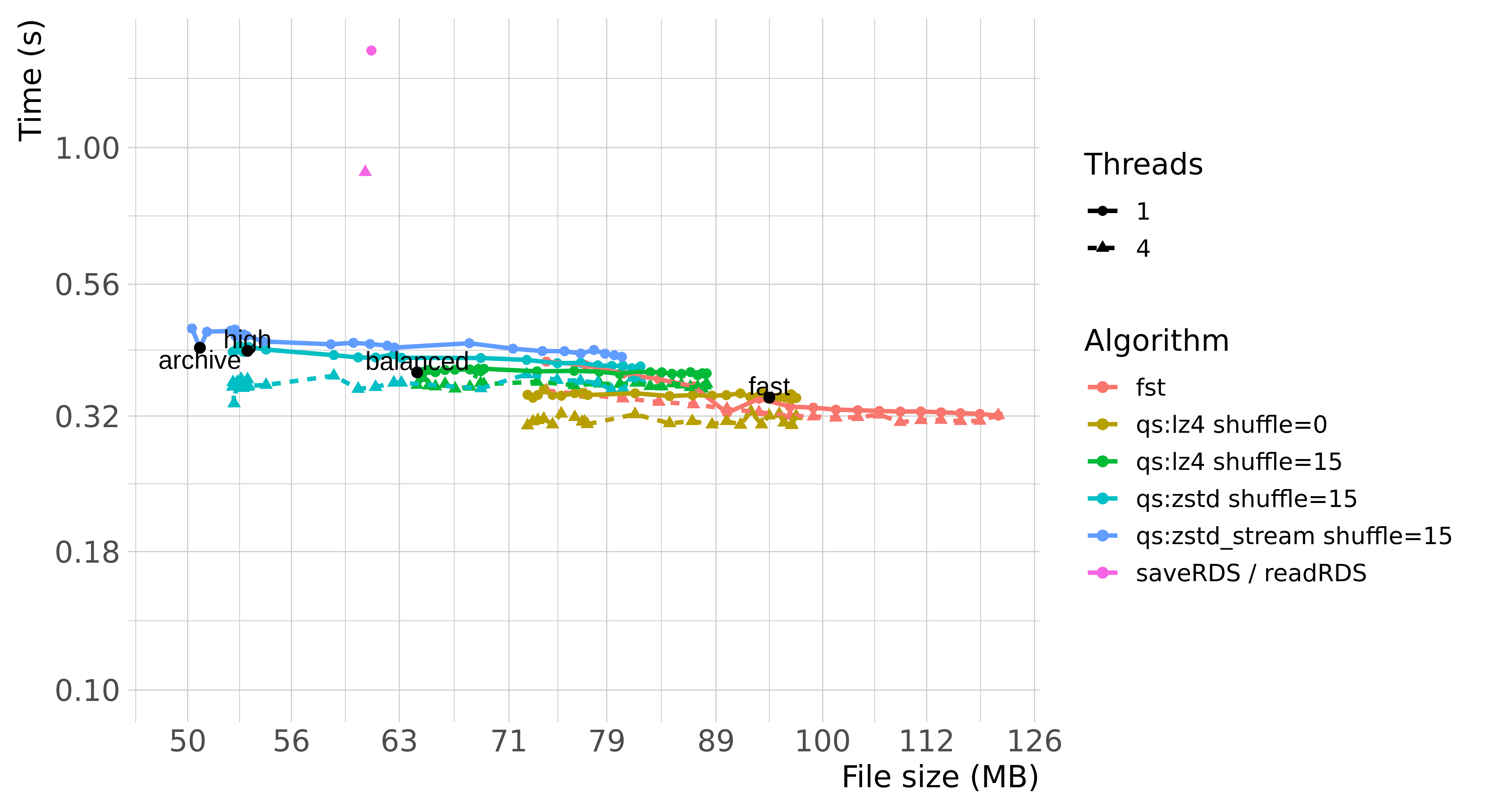
(Benchmarks are based on qs ver. 0.21.2,
fst ver. 0.9.0 and R 3.6.1.)
Benchmarking write and read speed is a bit tricky and depends highly on a number of factors, such as operating system, the hardware being run on, the distribution of the data, or even the state of the R instance. Reading data is also further subjected to various hardware and software memory caches.
Generally speaking, qs and fst are
considerably faster than saveRDS regardless of using single
threaded or multi-threaded compression. qs also manages to
achieve superior compression ratio through various optimizations
(e.g. see “Byte Shuffle” section below).
The ALTREP system (new as of R 3.5.0) allows package developers to represent R objects using their own custom memory layout. This allows a potentially large speedup in processing certain types of data.
In qs, ALTREP character vectors are
implemented via the stringfish
package and can be used by setting use_alt_rep=TRUE in the
qread function. The benchmark below shows the time it takes
to qread several million random strings
(nchar = 80) with and without ALTREP.
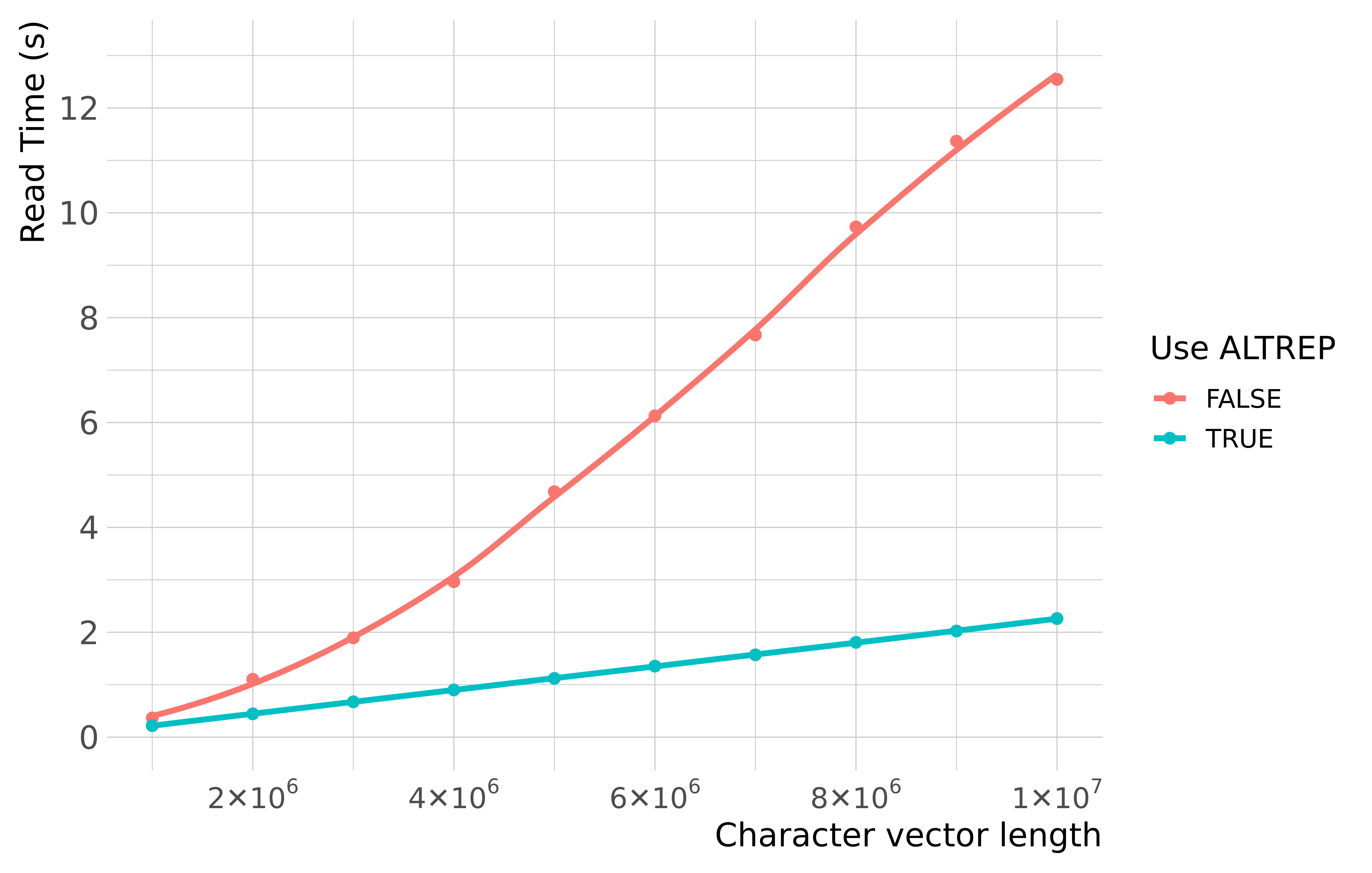
The large speedup demonstrates why one would want to consider the
system, but there are caveats. Downstream processing functions must be
ALTREP-aware. See the stringfish
package for more details.
Byte shuffling (adopted from the Blosc meta-compression library) is a way of re-organizing data to be more amenable to compression. An integer contains four bytes and the limits of an integer in R are +/- 2^31-1. However, most real data doesn’t use anywhere near the range of possible integer values. For example, if the data were representing percentages, 0% to 100%, the first three bytes would be unused and zero.
Byte shuffling rearranges the data such that all of the first bytes
are blocked together, all of the second bytes are blocked together, and
so on. This procedure often makes it very easy for compression
algorithms to find repeated patterns and can often improve compression
ratio by orders of magnitude. In the example below, shuffle compression
achieves a compression ratio of over 1:1000. See ?qsave for
more details.
# With byte shuffling
x <- 1:1e8
qsave(x, "mydat.qs", preset = "custom", shuffle_control = 15, algorithm = "zstd")
cat( "Compression Ratio: ", as.numeric(object.size(x)) / file.info("mydat.qs")$size, "\n" )
# Compression Ratio: 1389.164
# Without byte shuffling
x <- 1:1e8
qsave(x, "mydat.qs", preset = "custom", shuffle_control = 0, algorithm = "zstd")
cat( "Compression Ratio: ", as.numeric(object.size(x)) / file.info("mydat.qs")$size, "\n" )
# Compression Ratio: 1.479294 You can use qs to directly serialize objects to
memory.
Example:
library(qs)
x <- qserialize(c(1, 2, 3))
qdeserialize(x)
[1] 1 2 3The qs package includes two sets of utility functions
for converting binary data to ASCII:
base85_encode and base85_decodebase91_encode and base91_decodeThese functions are similar to base64 encoding functions found in various packages, but offer greater efficiency.
Example:
enc <- base91_encode(qserialize(datasets::mtcars, preset = "custom", compress_level = 22))
dec <- qdeserialize(base91_decode(enc))(Note: base91 strings contain double quote characters
(") and need to be single quoted if stored as a
string.)
See the help files for additional details and history behind these algorithms.
qs functions can be called directly within C++ code via
Rcpp.
Example C++ script:
// [[Rcpp::depends(qs)]]
#include <Rcpp.h>
#include <qs.h>
using namespace Rcpp;
// [[Rcpp::export]]
void test() {
qs::qsave(IntegerVector::create(1,2,3), "/tmp/myfile.qs", "high", "zstd", 1, 15, true, 1);
}R side:
library(qs)
library(Rcpp)
sourceCpp("test.cpp")
# save file using Rcpp interface
test()
# read in file created through Rcpp interface
qread("/tmp/myfile.qs")
[1] 1 2 3The C++ functions do not have default parameters; all parameters must be specified.
Future versions will be backwards compatible with the current version.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.