The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
Sercan Kahveci 2024-06-25
if(!require(remotes)){ install.packages("remotes") }
remotes::install_github("Spiritspeak/rapidsplit")The R package rapidsplithalf computes the permutated split-half reliability of reaction time tasks quickly and accurately, using a balanced splitting algorithm that ensures both halves are of maximally equal size regardless of how many subdivisions exist, for example due to conditions or stratification. As far as we know, this is the only R package for split-half reliability that does so. rapidsplit further allows for stratification of the splits, for example by stimulus, to ensure that the two halves are as similar to each other as possible. Kahveci et al. (2022) found that this stratification enhances the accuracy of the reliability estimate. Lastly, there is support for performing certain pre-processing operations after splitting the data rather than before.
library(rapidsplithalf)
data(foodAAT)
data(raceIAT)Let’s say we have a reaction time task with 2 conditions whose RTs we average and then subtract from each other within each participant. We want to know the reliability of this difference score. What do we do?
Kahveci et al. (2022) give a number of recommendations based on simulation analyses.
Split-half reliability far exceeds in precision compared to Cronbach’s alpha. They also find much better accuracy in performing many repeated random splits and averaging the result, rather than performing a single even-odd split.
Most common studies need up to 5500 splits to accurately detect the split-half reliability.
The accuracy of the reliability estimate is further improved by stratifying the random splits by stimulus such that both halves feature a near-equal number of trials for each stimulus, thereby ensuring that the two halves resemble replications of each other.
They recommend computing the reliability of the score that is actually used in analyses of the manuscript, i.e. if a difference score is used in correlational analyses, then the reliability of that difference score should be computed rather than of the means that were subtracted from each other. Likewise, if the data was aggregated using medians, then the reliability of a median-based score should be computed.
rel<-
rapidsplit(data=foodAAT,
subjvar="subjectid", # Subject identifier
diffvars="is_pull", # The variable indicating the two conditions that need to be subtracted from each other
stratvars="stimid", # splits are stratified by stimulus ID such that each half features a near-equal number of trials for each stimulus
aggvar="RT", # Defining the variable to be aggregated
aggfunc="means", # Method to aggregate each condition: simple means
splits=5500)## Generating splits...
## ================================================================================
## Aggregating conditions...
## ================================================================================print(rel)## Full-length reliability (Spearman-Brown coefficient):
## rSB (34) = .44, 95%CI [.04, .7], p = .007We can plot one of the splits from the reliability object to check for outliers.
plot(rel)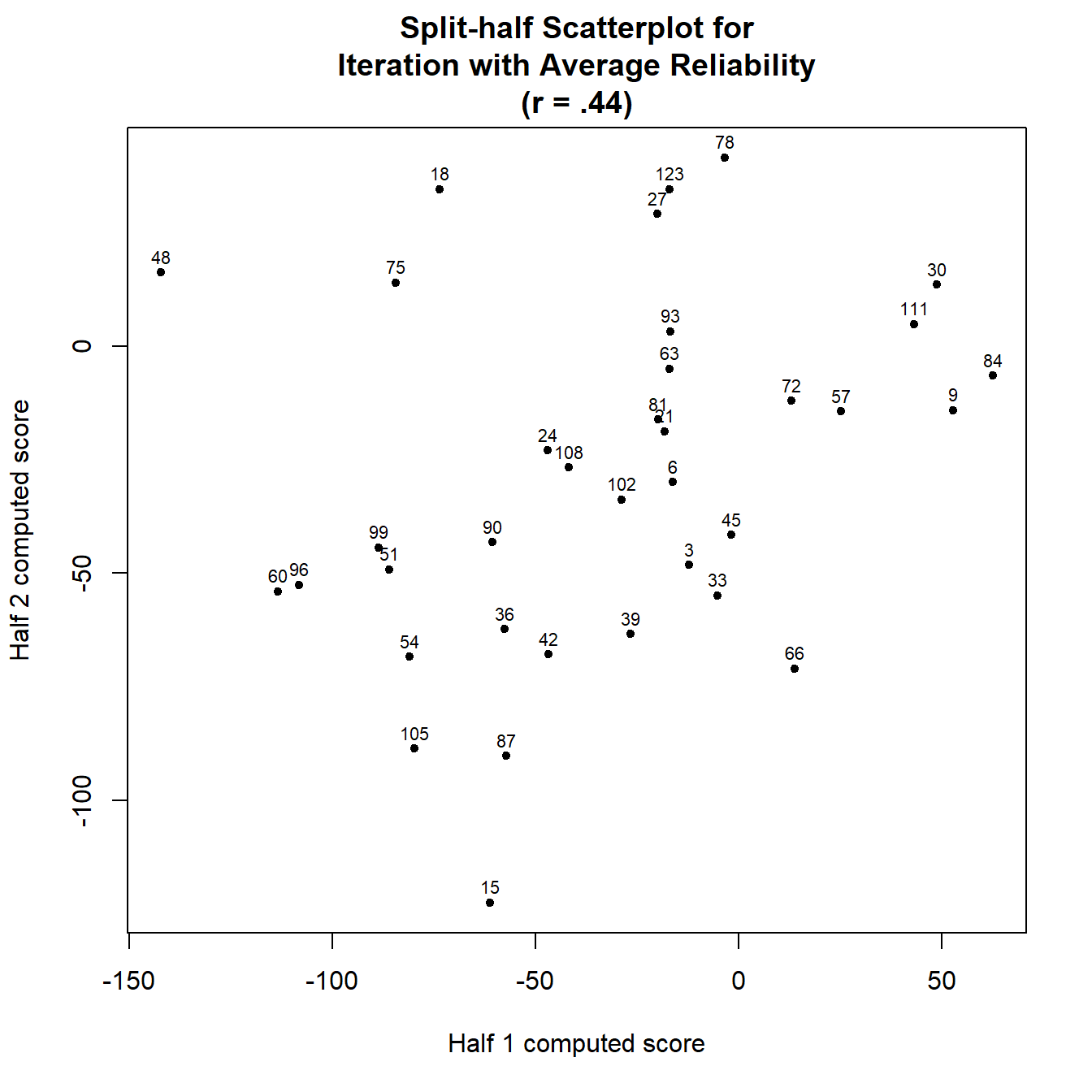
Or we can plot all of the splits simultaneously in a single figure.
plot(rel,type="all")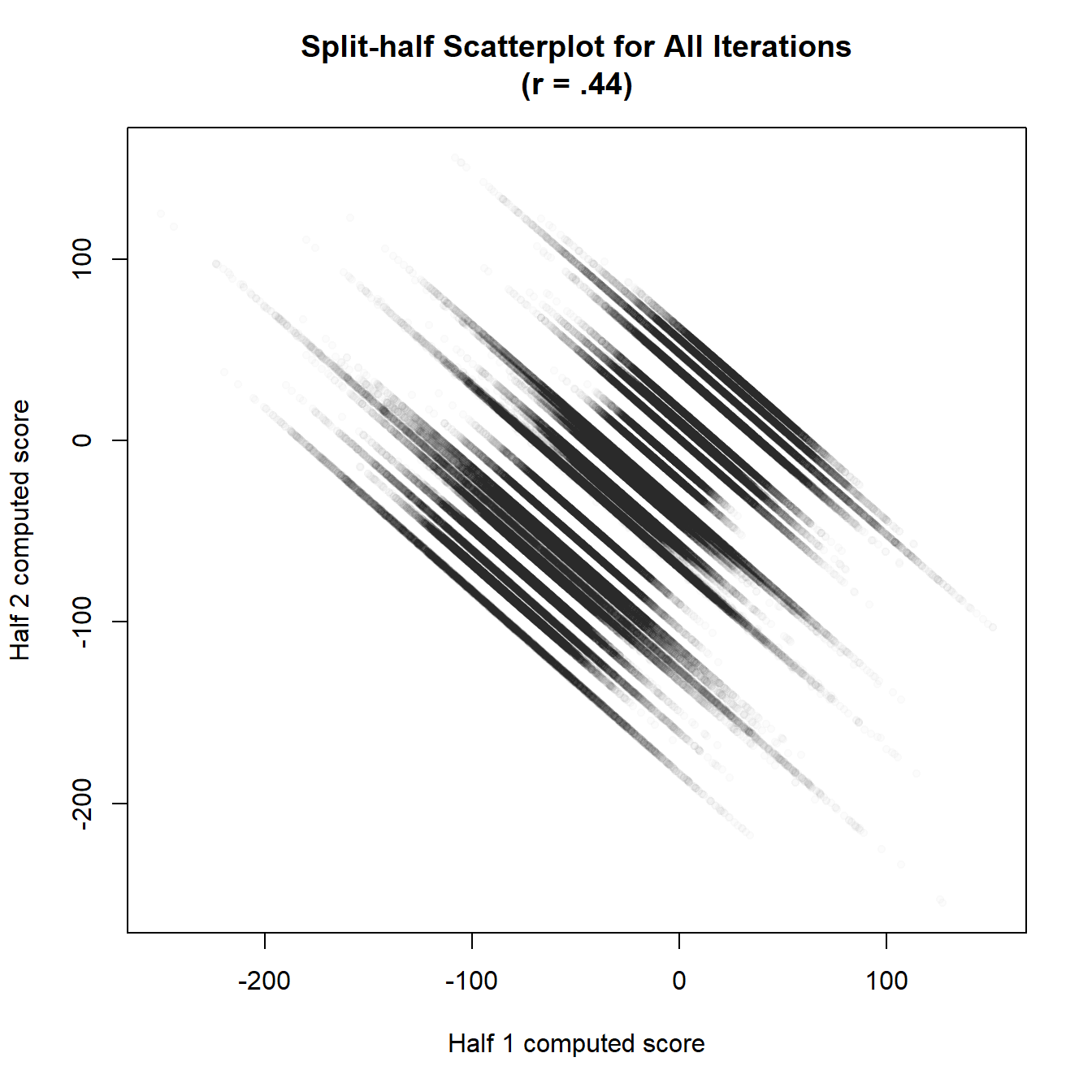
As you can see, the points fall into a diagonal line, with each line representing a single participant. The participants with a longer line have a more uncertain score.
As discussed, if you’re using medians to aggregate your conditions, then the reliability of a medin-based score should be computed. We do so here within the context of a double-difference score, common to the approach-avoidance task. This has the following format: (push target - pull target) - (push control - pull control)
rel2<-
rapidsplit(data=foodAAT,
subjvar="subjectid",
# We specify 2 diffvars for a double-difference score
diffvars=c("is_pull","is_target"),
stratvars="stimid",
aggvar="RT",
# We specify the median here
aggfunc="medians",
splits=5500)## Generating splits...
## ================================================================================
## Aggregating conditions...
## ================================================================================print(rel2)## Full-length reliability (Spearman-Brown coefficient):
## rSB (34) = .87, 95%CI [.79, .93], p = 0We can also compute the reliability of a D-score from an implicit association task.
iatrel<-
rapidsplit(data=raceIAT,
subjvar="session_id",
diffvars="congruent",
# The subscorevar argument specifies that we want to compute
# multiple scores for each participant, based on subsets of
# their data, and then average the scores together
# as is done in the IAT D-score.
subscorevar="blocktype",
aggvar="latency",
splits=1000,
# the errorhandling argument controls how error trials
# are replaced with the block mean plus a penalty,
# as is done in the IAT D-score.
errorhandling=list(type="fixedpenalty",
errorvar="error",
fixedpenalty=600,
blockvar="block_number"),
# The standardize argument specifies that we want to
# divide the person's score by the standard deviation of their
# RTs, as in the IAT D-score.
standardize=TRUE)## Generating splits...
## ================================================================================
## Replacing error trials
## ================================================================================
## Aggregating conditions...
## ================================================================================
## Computing standard deviations to standardize scores by...
## ================================================================================print(iatrel)## Full-length reliability (Spearman-Brown coefficient):
## rSB (198) = .84, 95%CI [.76, .89], p = 0plot(iatrel,show.labels=FALSE)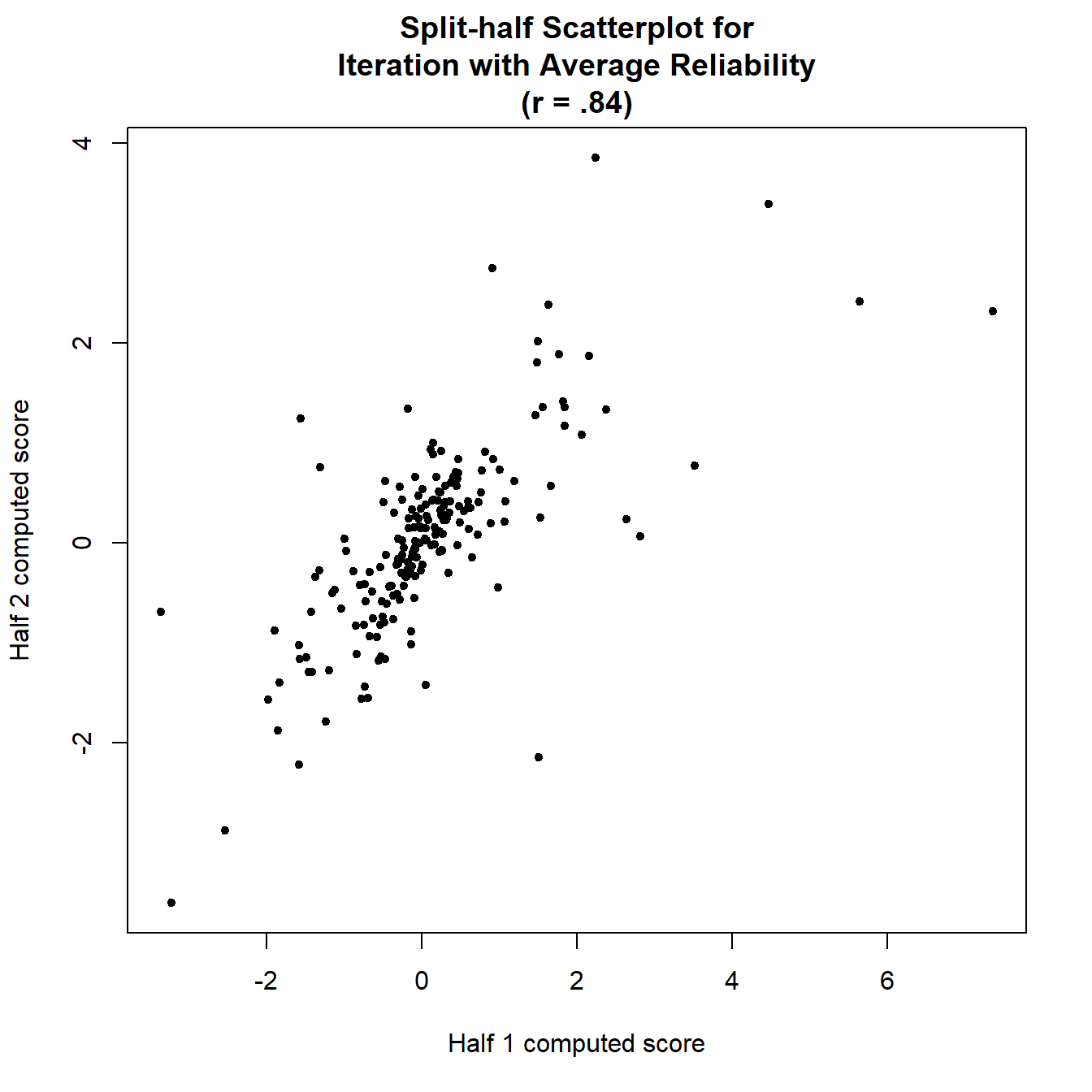
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.