
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
redcapAPI is an R package to pull data from a REDCap project. Its design
goes far beyond a ‘thin’ client which just exposes the raw REDCap API
into R. One principal goal is to get data into memory using base R in a
format that is analysis ready with a minimum of function calls. There
are over 7,000 institutions and 3 million users of REDCap worldwide
collecting data. Analysis in R for monitoring and reporting that data is
a common concern for these projects.
Core concerns handled by the library:
unlockREDCap. There are override methods available for
production environments.reviewInvalidRecords provides a summary report of all data
that fails validation, with hot links to the record in question. This is
an important step. Data that does not match the target format cannot be
cast, e.g. “xyz” cannot be treated as a numeric and will become NA in
the final data set.| Feature | redcapAPI | REDCapR | REDCapExporter | tidyREDCap | REDCapTidieR | REDCapDM | redquack |
|---|---|---|---|---|---|---|---|
| CRAN Downloads | |||||||
| Export Data To R | ✅ | ✅ | ✅ | ✅ | ✅ | ✅ | ✅ |
| Import Data From R | ✅ | ✅ | ❌ | ❌ | ❌ | ❌ | ❌ |
| Sparse Block Splitting | ✅ | ✅ | ❌ | ✅ | ✅ | ✅ | ❌ |
| Field Labeling | ✅ | ❌ | ❌ | ✅ | ❌ | ✅ | ❌ |
| Attribute Processing | ✅ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ |
| Logical Expression Query | partial | ❌ | ❌ | ❌ | ❌ | ✅ | ❌ |
| Tidy/Tibble Support | ❌ | ❌ | ❌ | ❌ | ✅ | ❌ | ✅ |
| Data Summary | ❌ | ❌ | ❌ | ❌ | ✅ | ❌ | ❌ |
| Type Conversion Callbacks | ✅ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ |
| API Failure Auto-Retry | ✅ | ❌ | ❌ | ❌ | ❌ | ❌ | ✅ |
| Secure API Key Storage | ✅ | ❌ | ✅ | ❌ | ❌ | ❌ | ❌ |
| Validation Reporting | ✅ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ |
| Extensive Test Suite | ✅ | ✅ | ❌ | ❌ | ✅ | ❌ | ✅ |
| Logfile Processing | ✅ | ❌ | ❌ | ❌ | ❌ | ❌ | ✅ |
| Offline Calculated Fields | ❌ | ❌ | ❌ | ❌ | ❌ | ✅ | ❌ |
There are 2 basic functions that are key to understanding the core approach:
unlockREDCapexportBulkRecordsHere’s a typical call for these two:
library(redcapAPI)
unlockREDCap(c(rcon = '<MY PROJECT NAME>'),
keyring = 'API_KEYs',
envir = globalenv(),
url = 'https://<REDCAP_URL>/api/')
exportBulkRecords(list(db = rcon),
forms = list(db = unique(rcon$metadata()$form_name)),
envir = globalenv())The <MY PROJECT NAME> is a reference for whatever
you wish to call this REDCap project. The rcon is the
variable you wish to assign it too. The keyring is a name for this key
ring. If one uses 'API_KEYs' for all your projects, you’ll
have one big keyring for all your API_KEYs locally encrypted. The url is
the standard url for the api at your institution. The envir
call is where to write the connection object; if not specified the call
will return a list.
The next call to exportBulkRecords, says to export by
form and leave out records not filled out and columns not part of a
form. The first argument is specifying a db reference to
the connection opened and naming it the same thing. The second call is
saying for this connection export back the all the forms/instruments
present in that db, if this is left blank it defaults to
all forms/instruments. The envir has it writing it back to
the global environment as variables. Any parameter not recognized is
passed to the exportRecordsTyped call–for every REDCap
database connection. For most analysis projects the function
exportBulkRecords provides the functionality required to
get the data in memory, converted, type cast and sparse block matrix
split into forms/instruments with blank rows filtered out.
These two calls will handle most analysis requests. To truly
understand all these changes see:
vignette("redcapAPI-best-practices").
2.7.0 introduced exportRecordsTyped which is a major
move forward for the package. It replaces exportRecords
with a far more stable and dependable call. It includes retries with
exponential back off through the connection object. It has inversion of
control over casting, and has a useful validation report attached when
things fail. It is worth the time to convert calls to
exportRecords to exportRecordsTyped and begin
using this new routine. It is planned that in the next year
exportRecords will be removed from the package.
This package exists to serve the research community and would not exist without community support. We are interested in volunteers who would like to translate the documentation into other languages.
If you wish to contribute new features to this software, we are open to pull requests. Before doing a lot of work, it would be best to open issue for discussion about your idea.
REDCap and it’s API have a large number of options and choices, with such complexity the possibility of bugs increases as well. This is a checklist for troubleshooting exports.
Rec <- exportRecordsTyped(rcon) give you a
warning about data that failed validations? If so, what kind of content
are you seeing from reviewInvalidRecords(Rec)?exportRecordsTyped(rcon, validation = skip_validation, cast = raw_cast)?
This is a completely raw export with no processing by the library.rcon$projectInformation()$missing_data_codesrcon$projectInformation()$secondary_unique_field. In
earlier versions REDCap will report one even if it’s been disabled
later, if this column doesn’t exist then the library is unable to
properly handle exports as the definition of the unique key doesn’t
exist. If one is defined and the field doesn’t exist, one will have to
contact their REDCap administrator to get the project fixed.filter_empty_rows=FALSE and see if that fixes it.packageVersion('redcapAPI').This means that the data/meta-data stored in the REDCap database contains improperly encoded characters. It is a problem with the REDCap project itself. The authors of this library do not know the root cause of the encoding issue, but suspect it was an earlier version of REDCap that did not handle encoding properly. This library is respecting the reported encoding type when loading into memory. All cases seen to date have the data encoded in ISO-8859-1 (the default when the HTTP header is missing charset) and the REDCap server treats all data as UTF-8. This improper coding can result in data loss via the GUI if records are updated. It is best to discuss with your institutions REDCap administrator how to repair this problem and such repairs are outside the scope of this library. This error message is to make one aware of this issue in their project. The library does the best it can when it encounters encoding issues.
If you need help or assistance in understanding how to approach a project or problem using the library, please open an issue. We use these questions to refine the documentation. Thus asking questions contributes to refinement of documentation.
Your institutions installation of REDCap contains a lot of documentation for the general usage of REDCap. For general questions outside the scope of interfacing the API to R please refer to your institutions REDCap instance documentation.
The help pages for functions is fairly extensive. Try
?exportRecordsTyped or
?fieldValidationAndCasting for good starting points into
the help pages.
There are several vignettes with helpful information and examples to explore. These provide higher level views than can be provided in help pages.
We strive to keep the dependency chain as minimal as possible. This reduces potential breakage due to downstream packages getting changed and minimizes install time.
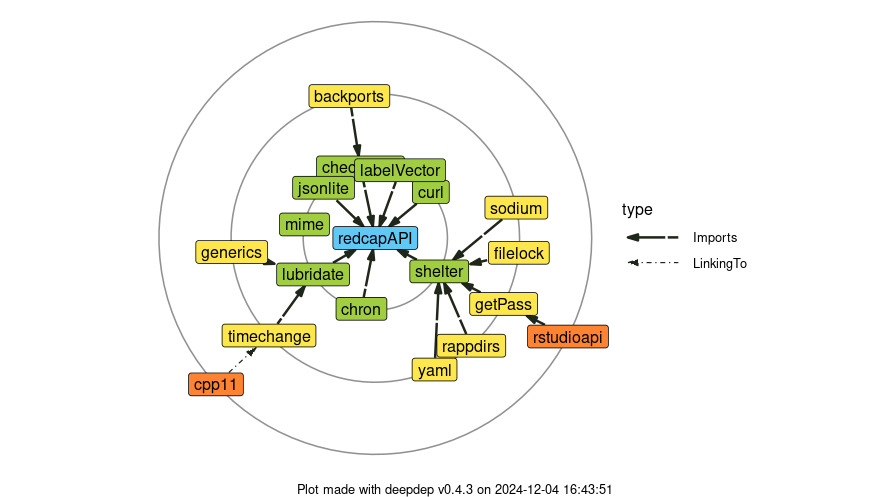
NOTE: Ownership transfer of this package to VUMC Biostatistics is complete.
The research community owes a big thanks to Benjamin Nutter for his years of service keeping this package current.
This package was originally created by Jeffrey Horner.
The current package was developed under REDCap Version 14+. Institutions can be a little behind on updating REDCap and so some features of the API may not always work.
Goals:
Rules:
redcapAPI A rich API client for interfacing REDCap to R
Copyright (C) 2012 Jeffrey Horner, Vanderbilt University
Copyright (C) 2013-2022 Benjamin Nutter
Copyright (C) 2023-2025 Benjamin Nutter, Shawn Garbett, Vanderbilt University
This program is free software; you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation; either version 2 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program; if not, write to the Free Software Foundation, Inc., 51 Franklin Street, Fifth Floor, Boston, MA 02110-1301, USA.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.