The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
The goal of scUtils is to collect utility functions that make single-cell RNAseq data analysis simple and understandable for anyone. At the same time, I will use it when writing my PhD thesis.
You can install the released version of scUtils from CRAN with:
install.packages("scUtils")And the development version from GitHub with:
# install.packages("devtools")
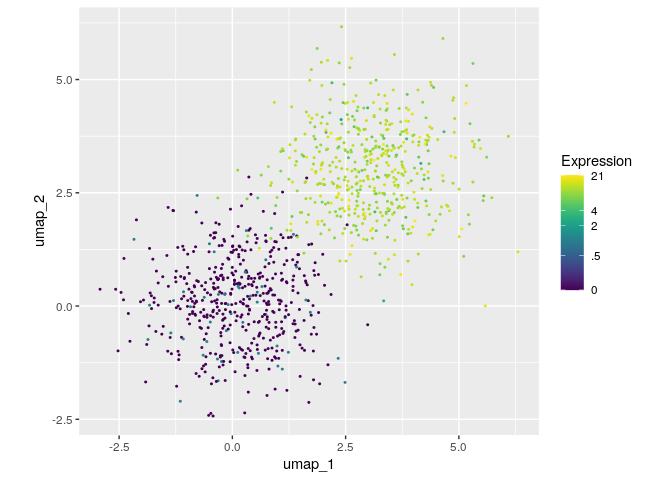
devtools::install_github("FelixTheStudent/scUtils")Feature Plots highlight gene expression in a 2-dimensional embedding (computed e.g. with UMAP or tSNE).
library(scUtils)
# simulate some data
set.seed(100)
my_umap <- matrix(rnorm(2000, c(.1, 3)), ncol=2, dimnames = list(NULL, c("umap_1", "umap_2")))
my_expr <- rpois(1000, c(.1, 11))
feat(my_umap, my_expr)
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.