
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
This package offers a very bare-bones interface to use the Metropolis-Hastings Monte Carlo Markov Chain algorithm. It is suitable for teaching and testing purposes. For more advanced uses, you can check out the mcmcensemble or adaptMCMC packages, which are designed with a very similar interface, but often allow better convergence, especially for badly scaled problems or highly correlated set of parameters.
You can install this package from CRAN:
install.packages("simpleMH")or from my r-universe (development version):
install.packages("simpleMH", repos = "https://bisaloo.r-universe.dev")library(simpleMH)
## a log-pdf to sample from
p.log <- function(x) {
B <- 0.03 # controls 'bananacity'
-x[1]^2/200 - 1/2*(x[2]+B*x[1]^2-100*B)^2
}
res <- simpleMH(
p.log,
inits = c(0, 0),
theta.cov = diag(2),
max.iter = 5000,
coda = TRUE # to be able to have nice plots and diagnostics with the coda pkg
)Here is the resulting sampling landscape of p.log():
plot(as.data.frame(res$samples))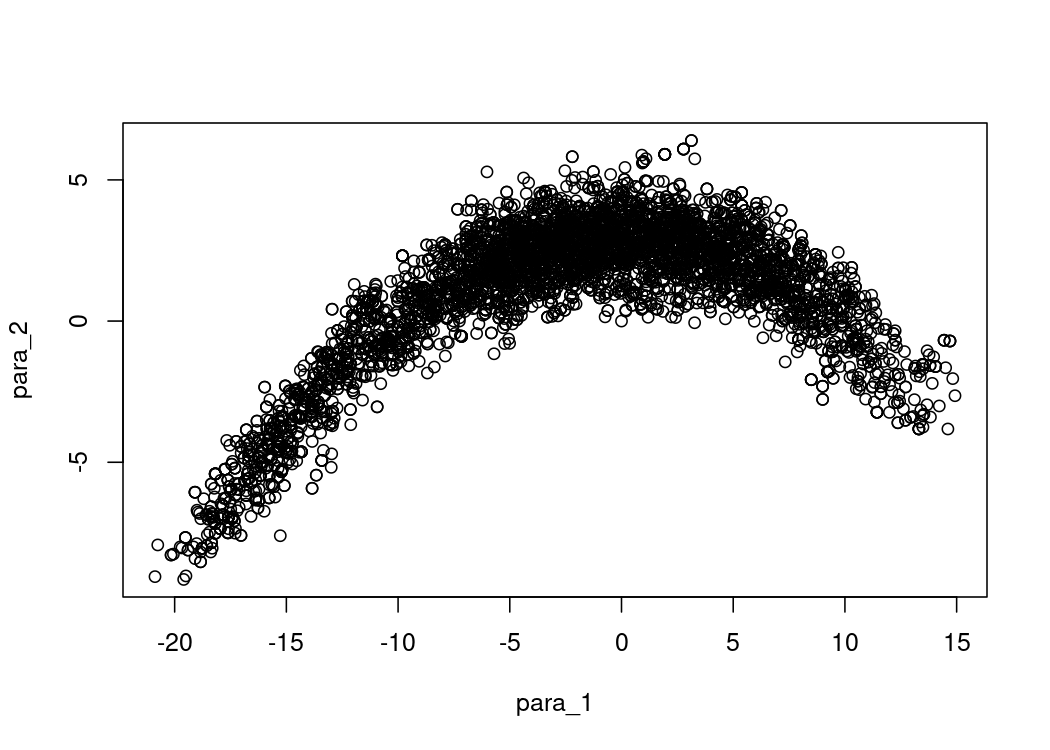
We can then use the coda package to post-process the chain (burn-in, thinning, etc.), plot the trace and density, or compute convergence diagnostics:
plot(res$samples)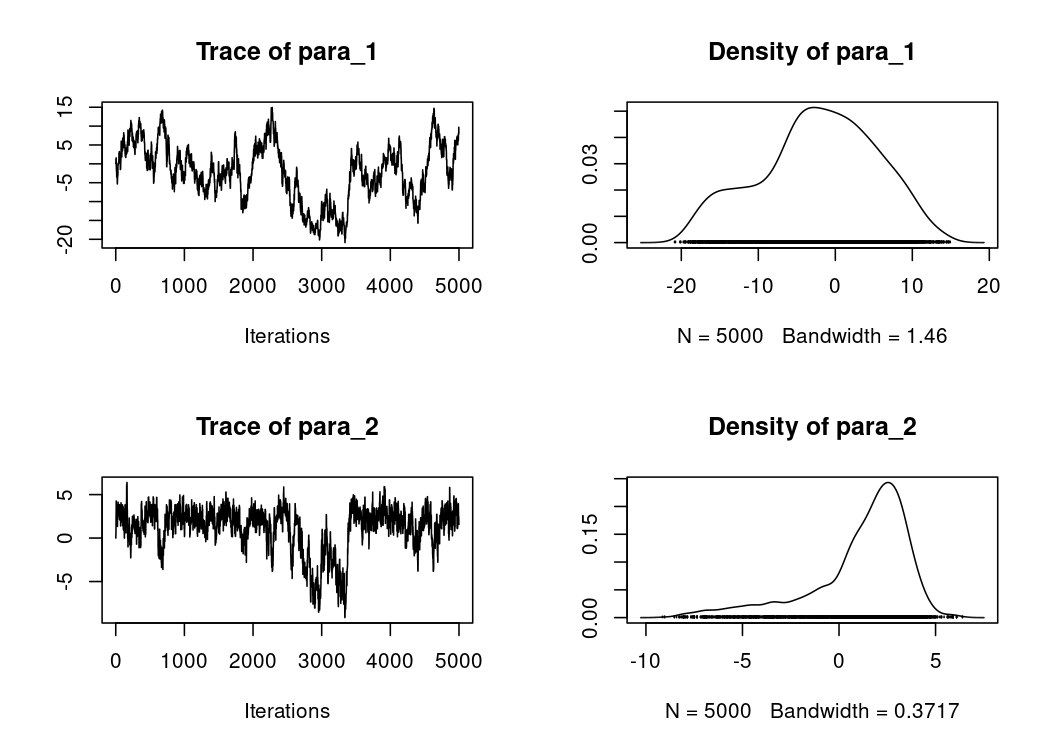
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.