
The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
{statsExpressions}:
Tidy dataframes and expressions with statistical details| Status | Usage | Miscellaneous |
|---|---|---|
 |
The {statsExpressions} package has two key aims:
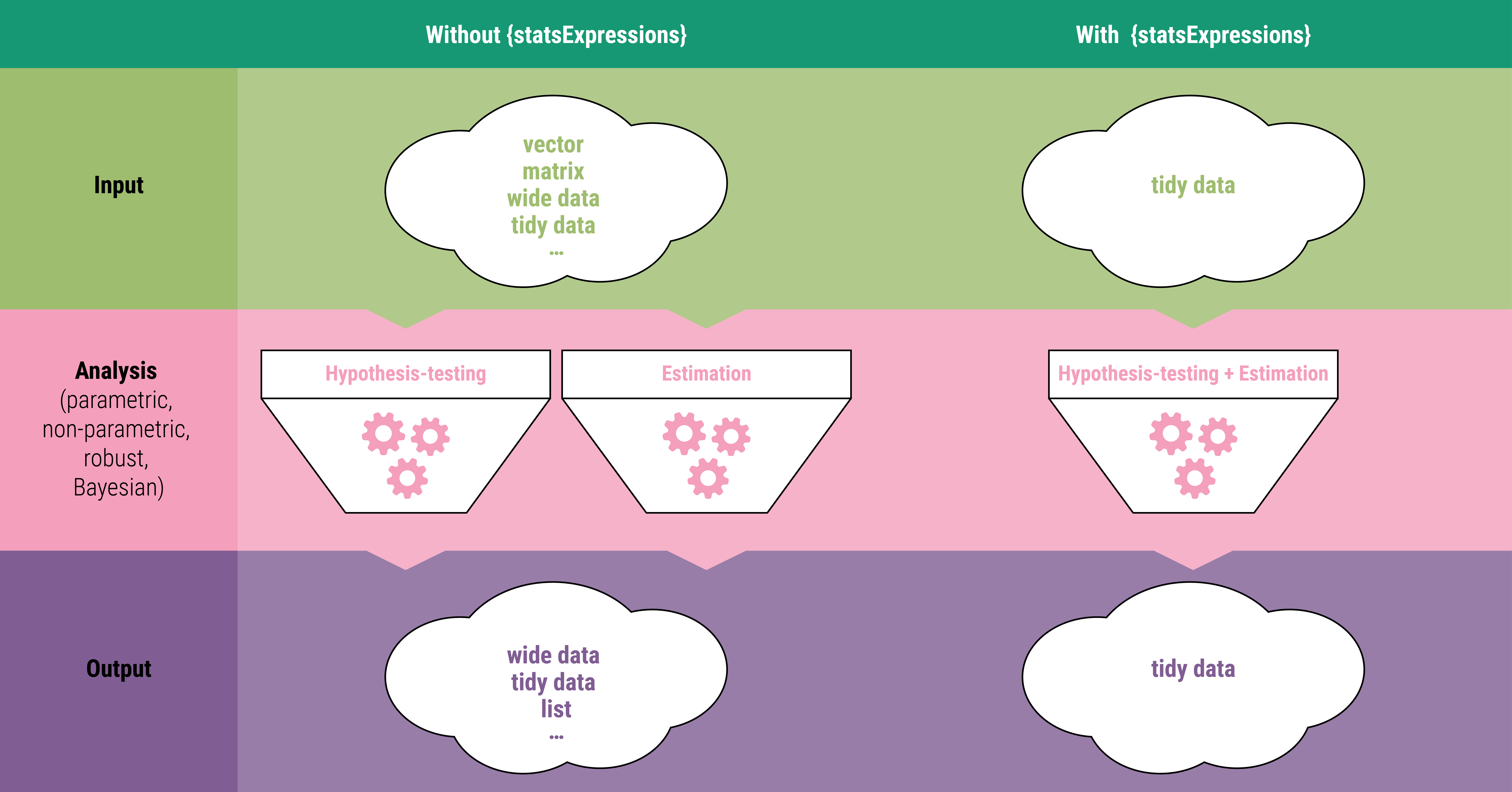
Statistical packages exhibit substantial diversity in terms of their syntax and expected input type. This can make it difficult to switch from one statistical approach to another. For example, some functions expect vectors as inputs, while others expect dataframes. Depending on whether it is a repeated measures design or not, different functions might expect data to be in wide or long format. Some functions can internally omit missing values, while other functions error in their presence. Furthermore, if someone wishes to utilize the objects returned by these packages downstream in their workflow, this is not straightforward either because even functions from the same package can return a list, a matrix, an array, a dataframe, etc., depending on the function.
This is where {statsExpressions} comes in: It can be
thought of as a unified portal through which most of the functionality
in these underlying packages can be accessed, with a simpler interface
and no requirement to change data format.
This package forms the statistical processing backend for ggstatsplot
package.
For more documentation, see the dedicated website.
| Type | Command |
|---|---|
| Release | install.packages("statsExpressions") |
| Development | pak::pak("IndrajeetPatil/statsExpressions") |
On Linux, {statsExpressions} installation may require
additional system dependencies, which can be checked using:
pak::pkg_sysreqs("statsExpressions")The package can be cited as:
citation("statsExpressions")
To cite package 'statsExpressions' in publications use:
Patil, I., (2021). statsExpressions: R Package for Tidy Dataframes
and Expressions with Statistical Details. Journal of Open Source
Software, 6(61), 3236, https://doi.org/10.21105/joss.03236
A BibTeX entry for LaTeX users is
@Article{,
doi = {10.21105/joss.03236},
year = {2021},
publisher = {{The Open Journal}},
volume = {6},
number = {61},
pages = {3236},
author = {Indrajeet Patil},
title = {{statsExpressions: {R} Package for Tidy Dataframes and Expressions with Statistical Details}},
journal = {{Journal of Open Source Software}},
}
Summary of available analyses
| Test | Function |
|---|---|
| one-sample t-test | one_sample_test() |
| two-sample t-test | two_sample_test() |
| one-way ANOVA | oneway_anova() |
| correlation analysis | corr_test() |
| contingency table analysis | contingency_table() |
| meta-analysis | meta_analysis() |
| pairwise comparisons | pairwise_comparisons() |
Summary of details available for analyses
| Analysis | Hypothesis testing | Effect size estimation |
|---|---|---|
| (one/two-sample) t-test | ✅ | ✅ |
| one-way ANOVA | ✅ | ✅ |
| correlation | ✅ | ✅ |
| (one/two-way) contingency table | ✅ | ✅ |
| random-effects meta-analysis | ✅ | ✅ |
Summary of supported statistical approaches
| Description | Parametric | Non-parametric | Robust | Bayesian |
|---|---|---|---|---|
| Between group/condition comparisons | ✅ | ✅ | ✅ | ✅ |
| Within group/condition comparisons | ✅ | ✅ | ✅ | ✅ |
| Distribution of a numeric variable | ✅ | ✅ | ✅ | ✅ |
| Correlation between two variables | ✅ | ✅ | ✅ | ✅ |
| Association between categorical variables | ✅ | ✅ | ❌ | ✅ |
| Equal proportions for categorical variable levels | ✅ | ✅ | ❌ | ✅ |
| Random-effects meta-analysis | ✅ | ❌ | ✅ | ✅ |
To illustrate the simplicity of this syntax, let’s say we want to run a one-way ANOVA. If we first run a non-parametric ANOVA and then decide to run a robust ANOVA instead, the syntax remains the same and the statistical approach can be modified by changing a single argument:
mtcars %>% oneway_anova(cyl, wt, type = "nonparametric")
#> # A tibble: 1 × 15
#> parameter1 parameter2 statistic df.error p.value
#> <chr> <chr> <dbl> <int> <dbl>
#> 1 wt cyl 22.8 2 0.0000112
#> method effectsize estimate conf.level conf.low
#> <chr> <chr> <dbl> <dbl> <dbl>
#> 1 Kruskal-Wallis rank sum test Epsilon2 (rank) 0.736 0.95 0.624
#> conf.high conf.method conf.iterations n.obs expression
#> <dbl> <chr> <int> <int> <list>
#> 1 1 percentile bootstrap 100 32 <language>
mtcars %>% oneway_anova(cyl, wt, type = "robust")
#> # A tibble: 1 × 12
#> statistic df df.error p.value
#> <dbl> <dbl> <dbl> <dbl>
#> 1 12.7 2 12.2 0.00102
#> method
#> <chr>
#> 1 A heteroscedastic one-way ANOVA for trimmed means
#> effectsize estimate conf.level conf.low conf.high
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Explanatory measure of effect size 1.05 0.95 0.843 1.50
#> n.obs expression
#> <int> <list>
#> 1 32 <language>All possible output dataframes from functions are tabulated here: https://indrajeetpatil.github.io/statsExpressions/articles/web_only/dataframe_outputs.html
Needless to say this will also work with the kable
function to generate a table:
set.seed(123)
# one-sample robust t-test
# we will leave `expression` column out; it's not needed for using only the dataframe
mtcars %>%
one_sample_test(wt, test.value = 3, type = "robust") %>%
dplyr::select(-expression) %>%
knitr::kable()| statistic | p.value | n.obs | method | effectsize | estimate | conf.level | conf.low | conf.high |
|---|---|---|---|---|---|---|---|---|
| 1.179181 | 0.275 | 32 | Bootstrap-t method for one-sample test | Trimmed mean | 3.197 | 0.95 | 2.854246 | 3.539754 |
These functions are also compatible with other popular data manipulation packages.
For example, let’s say we want to run a one-sample t-test
for all levels of a certain grouping variable. We can use
dplyr to do so:
# for reproducibility
set.seed(123)
library(dplyr)
# grouped operation
# running one-sample test for all levels of grouping variable `cyl`
mtcars %>%
group_by(cyl) %>%
group_modify(~ one_sample_test(.x, wt, test.value = 3), .keep = TRUE) %>%
ungroup()
#> # A tibble: 3 × 16
#> cyl mu statistic df.error p.value method alternative
#> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
#> 1 4 3 -4.16 10 0.00195 One Sample t-test two.sided
#> 2 6 3 0.870 6 0.418 One Sample t-test two.sided
#> 3 8 3 4.92 13 0.000278 One Sample t-test two.sided
#> effectsize estimate conf.level conf.low conf.high conf.method
#> <chr> <dbl> <dbl> <dbl> <dbl> <chr>
#> 1 Hedges' g -1.16 0.95 -1.88 -0.402 ncp
#> 2 Hedges' g 0.286 0.95 -0.388 0.937 ncp
#> 3 Hedges' g 1.24 0.95 0.544 1.91 ncp
#> conf.distribution n.obs expression
#> <chr> <int> <list>
#> 1 t 11 <language>
#> 2 t 7 <language>
#> 3 t 14 <language>Note that expression here means a pre-formatted
in-text statistical result. In addition to other details
contained in the dataframe, there is also a column titled
expression, which contains expression with statistical
details and can be displayed in a plot.
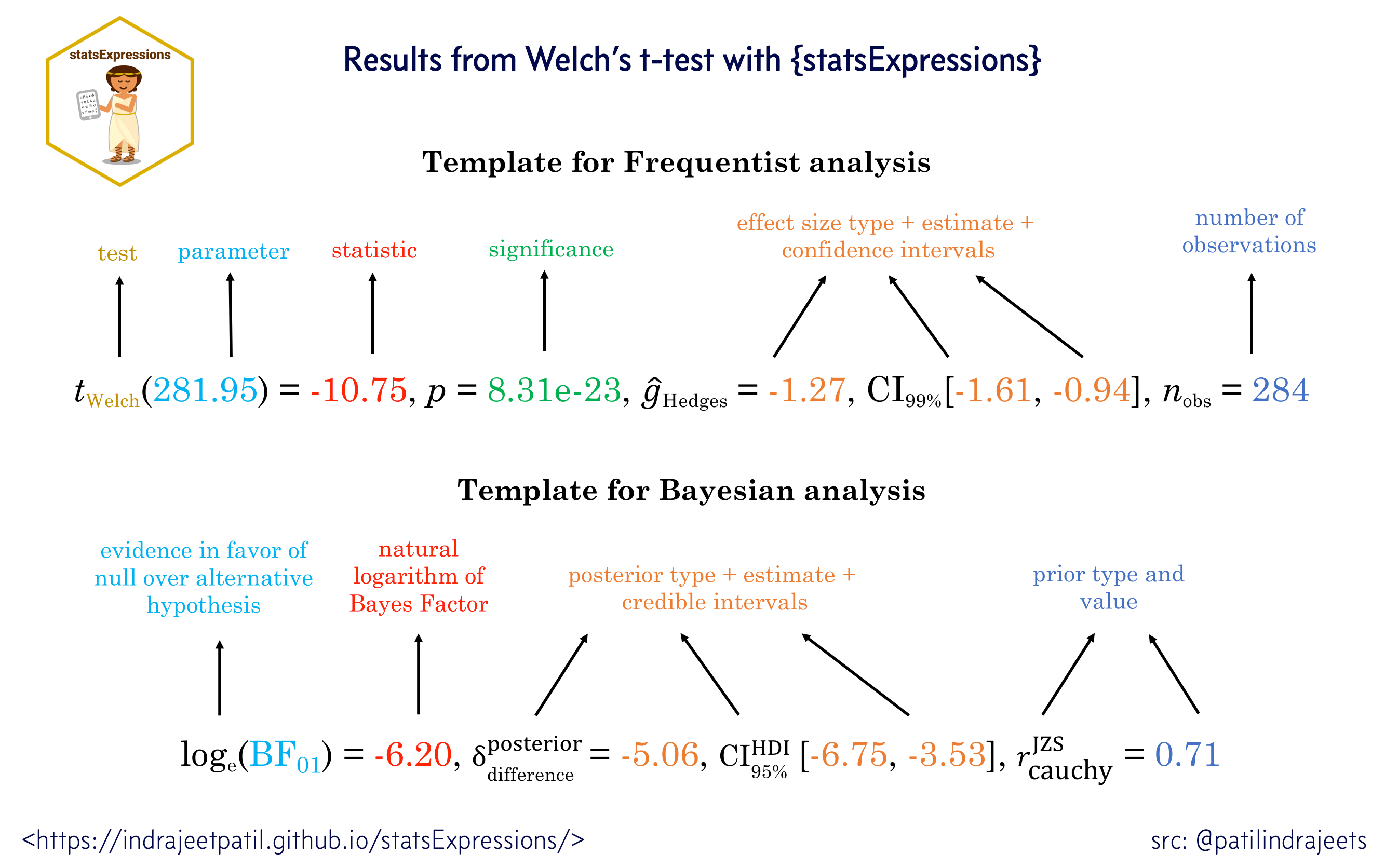
For all statistical test expressions, the default template attempt to follow the gold standard for statistical reporting.
For example, here are results from Welch’s t-test:
Let’s load the needed library for visualization:
library(ggplot2)Note that when used in a geometric layer, the expression need to be parsed.
# displaying mean for each level of `cyl`
centrality_description(mtcars, cyl, wt) |>
ggplot(aes(cyl, wt)) +
geom_point() +
geom_label(aes(label = expression), parse = TRUE)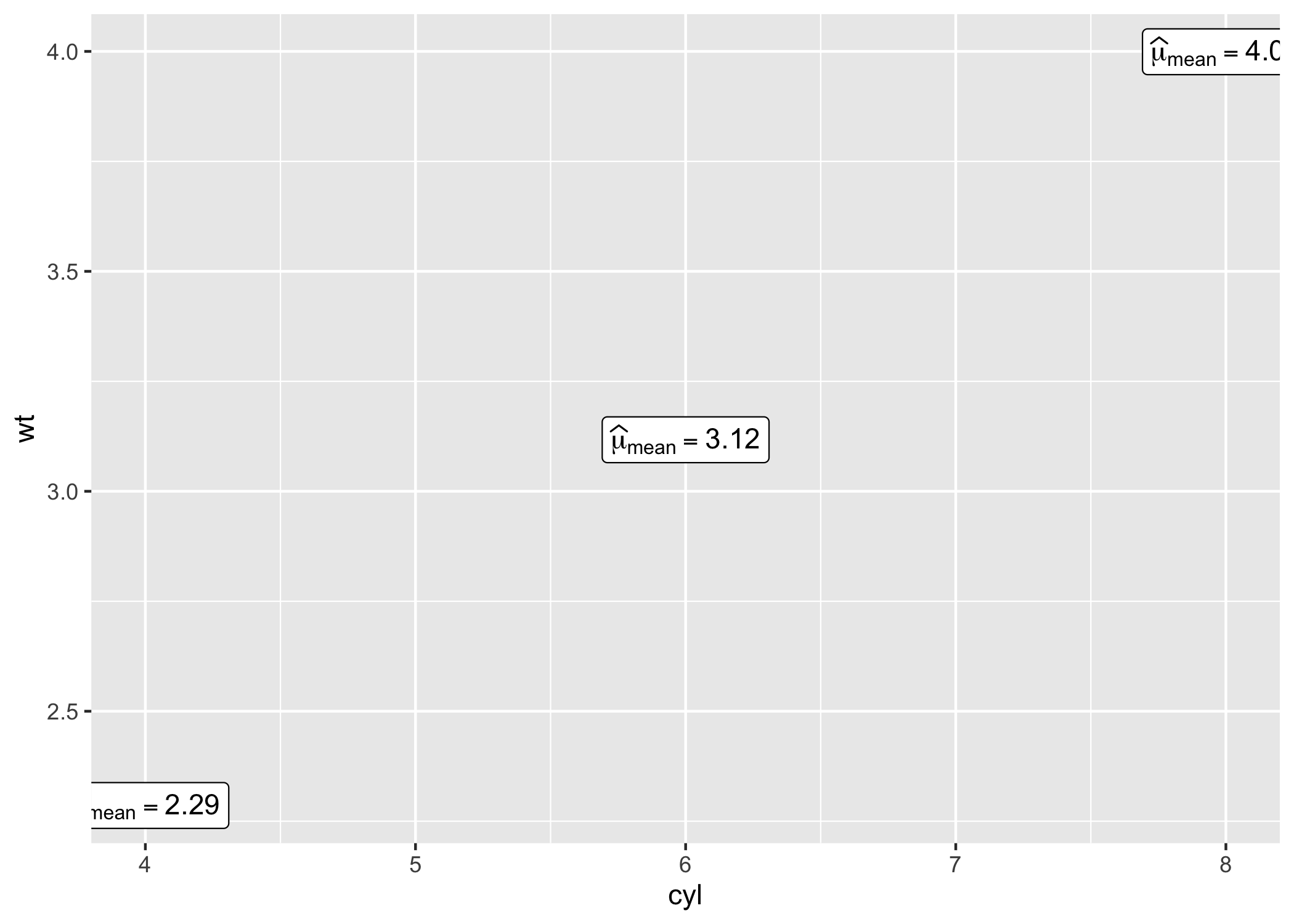
Here are a few examples for supported analyses.
The returned data frame will always have a column called
expression.
Assuming there is only a single result you need to display in a plot, to use it in a plot, you have two options:
results_data$expression[[1]]) without parsingparse(text = results_data$expression))If you want to display more than one expression in a plot, you will have to parse them.
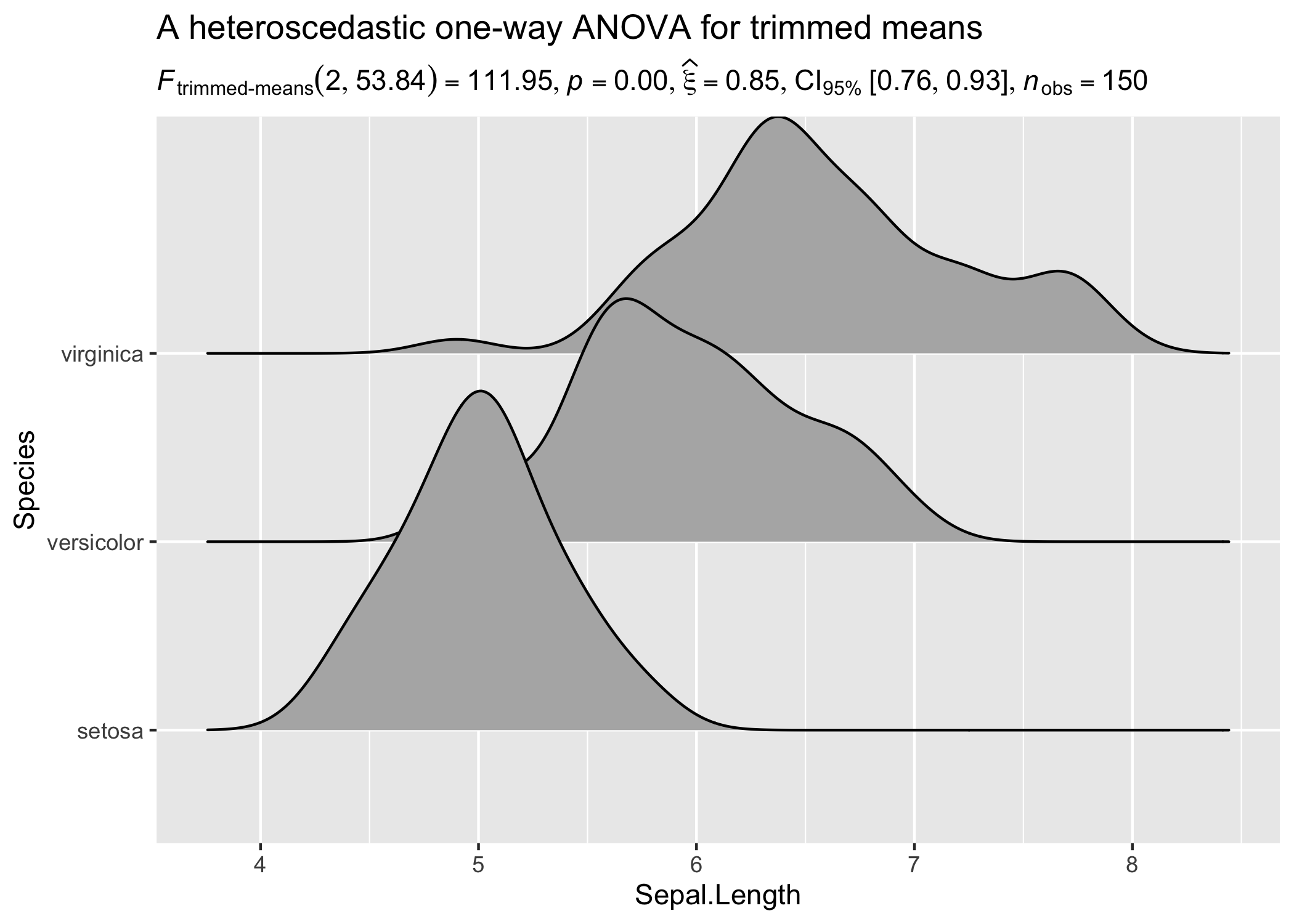
set.seed(123)
library(ggridges)
results_data <- oneway_anova(iris, Species, Sepal.Length, type = "robust")
# create a ridgeplot
ggplot(iris, aes(x = Sepal.Length, y = Species)) +
geom_density_ridges() +
labs(
title = "A heteroscedastic one-way ANOVA for trimmed means",
subtitle = results_data$expression[[1]]
)
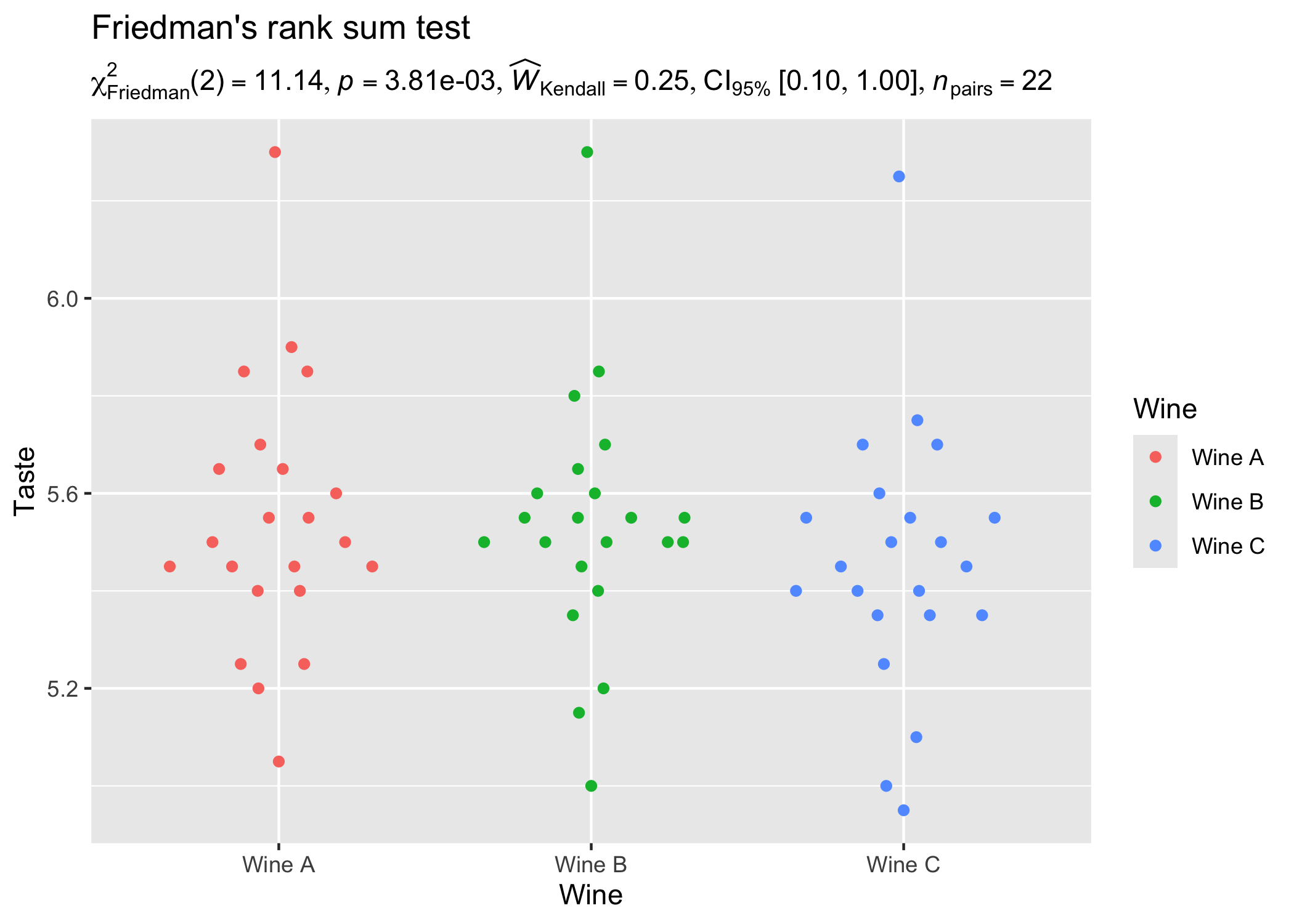
set.seed(123)
library(WRS2)
library(ggbeeswarm)
results_data <- oneway_anova(
WineTasting,
Wine,
Taste,
paired = TRUE,
subject.id = Taster,
type = "np"
)
ggplot2::ggplot(WineTasting, aes(Wine, Taste, color = Wine)) +
geom_quasirandom() +
labs(
title = "Friedman's rank sum test",
subtitle = parse(text = results_data$expression)
)
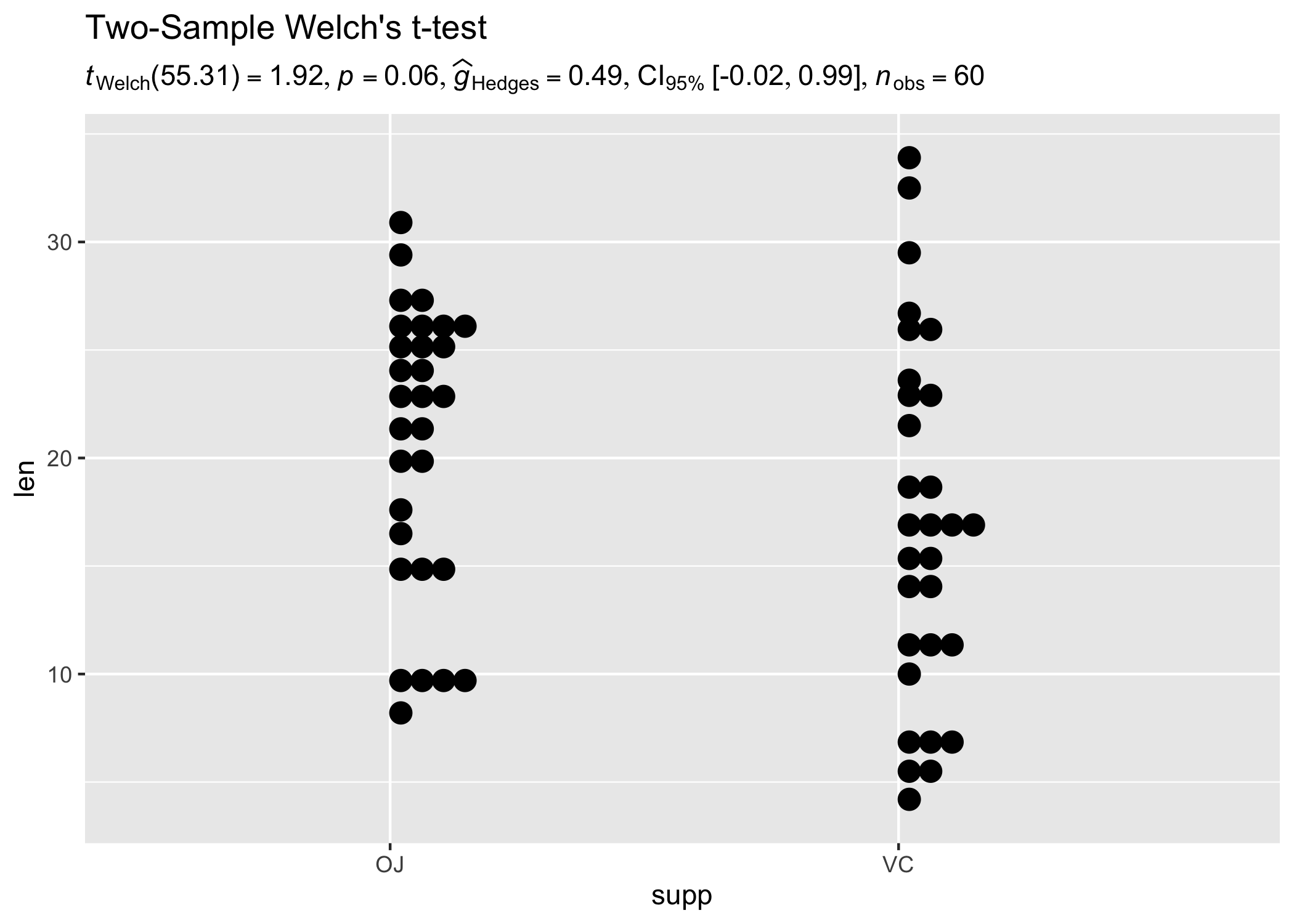
set.seed(123)
results_data <- two_sample_test(ToothGrowth, supp, len)
ggplot(ToothGrowth, aes(supp, len)) +
geom_boxplot() +
labs(
title = "Two-Sample Welch's t-test",
subtitle = parse(text = results_data$expression)
)
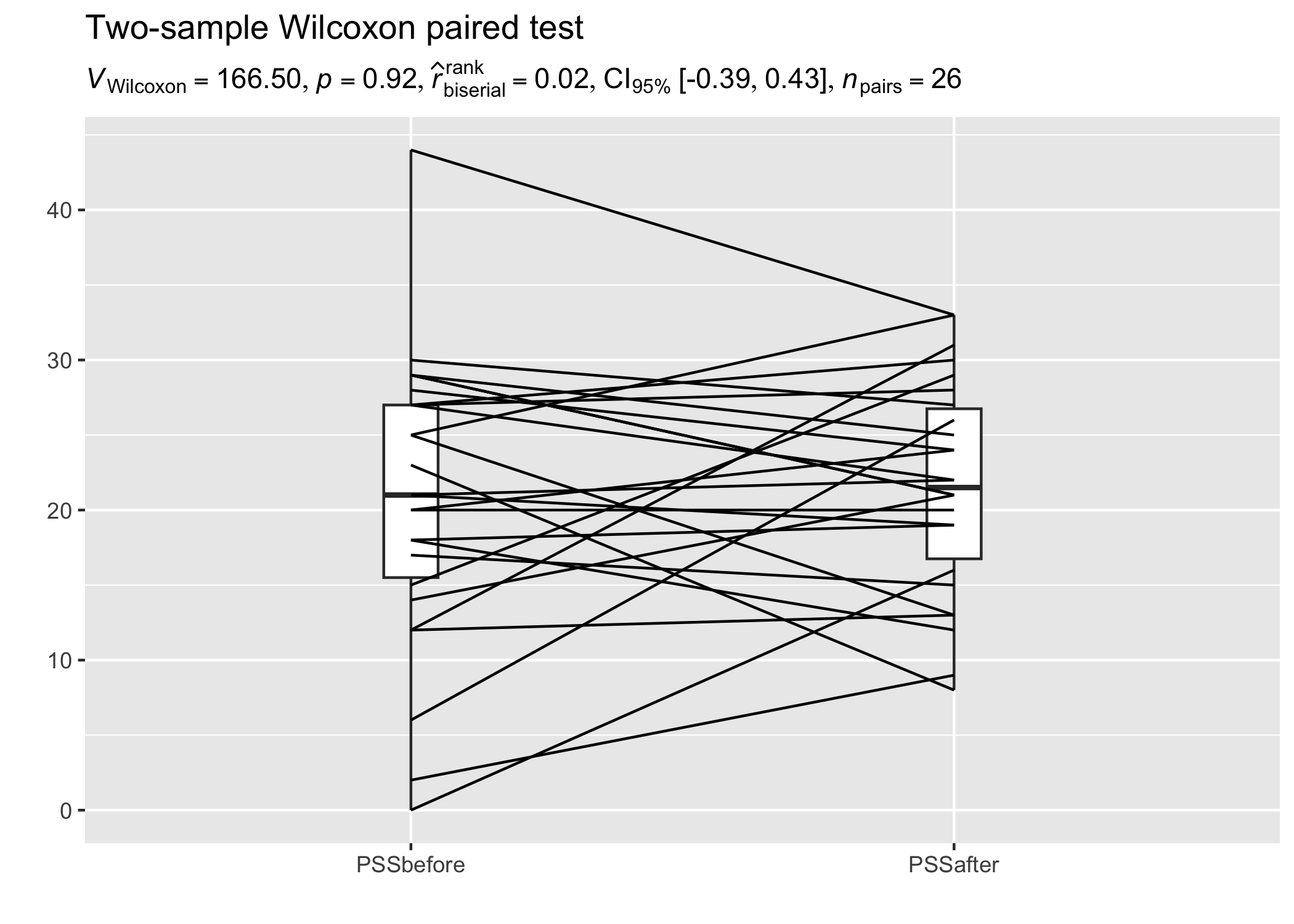
set.seed(123)
library(tidyr)
library(PairedData)
data(PrisonStress)
# get data in tidy format
df <- pivot_longer(PrisonStress, starts_with("PSS"), names_to = "PSS", values_to = "stress")
results_data <- two_sample_test(
data = df,
x = PSS,
y = stress,
paired = TRUE,
subject.id = Subject,
type = "np"
)
# plot
suppressWarnings(paired.plotProfiles(
PrisonStress, "PSSbefore", "PSSafter",
subjects = "Subject"
)) +
labs(
title = "Two-sample Wilcoxon paired test",
subtitle = parse(text = results_data$expression)
)
set.seed(123)
# dataframe with results
results_data <- one_sample_test(mtcars, wt, test.value = 3, type = "bayes")
# creating a histogram plot
ggplot(mtcars, aes(wt)) +
geom_histogram(alpha = 0.5) +
geom_vline(xintercept = mean(mtcars$wt), color = "red") +
labs(subtitle = parse(text = results_data$expression))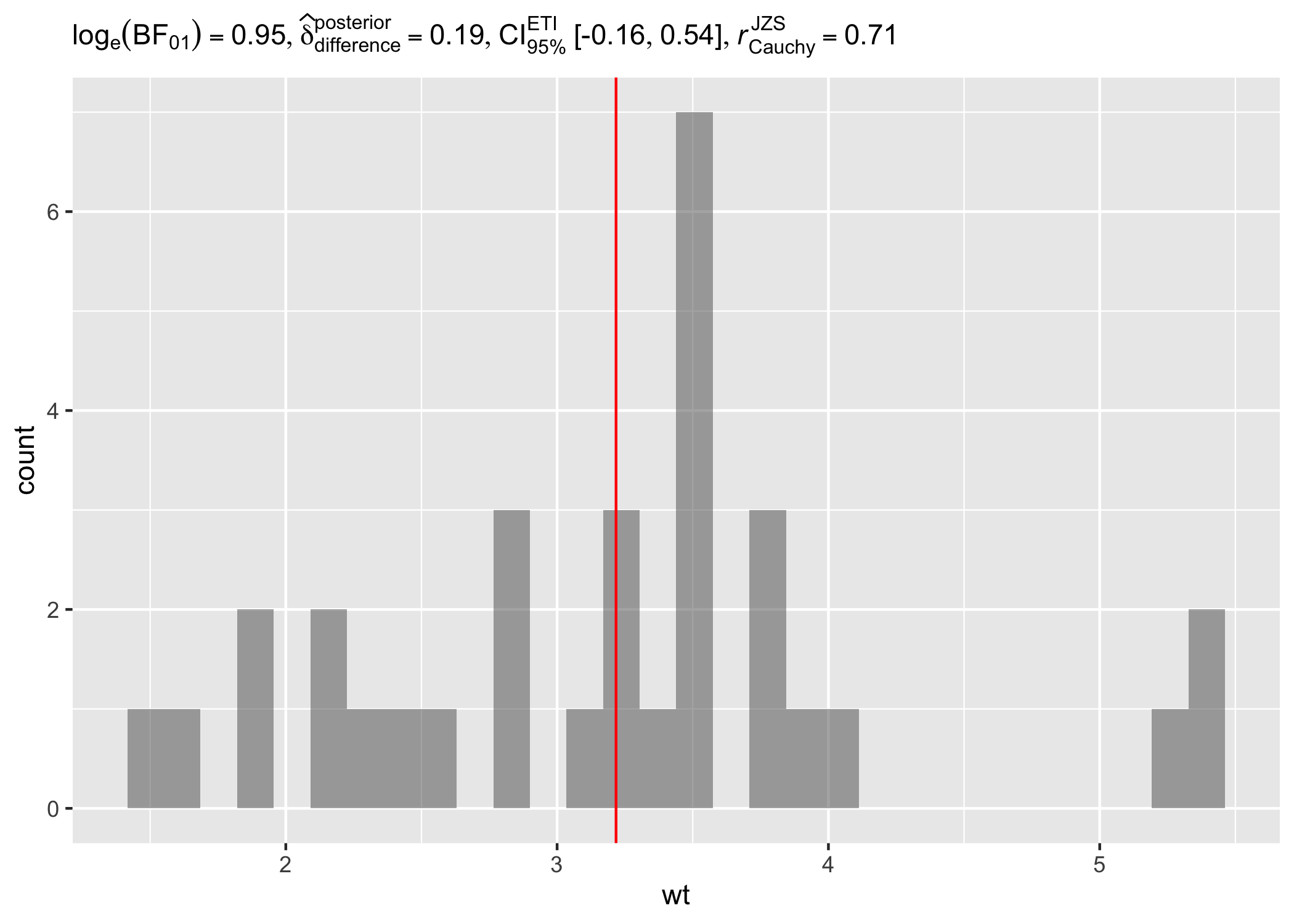
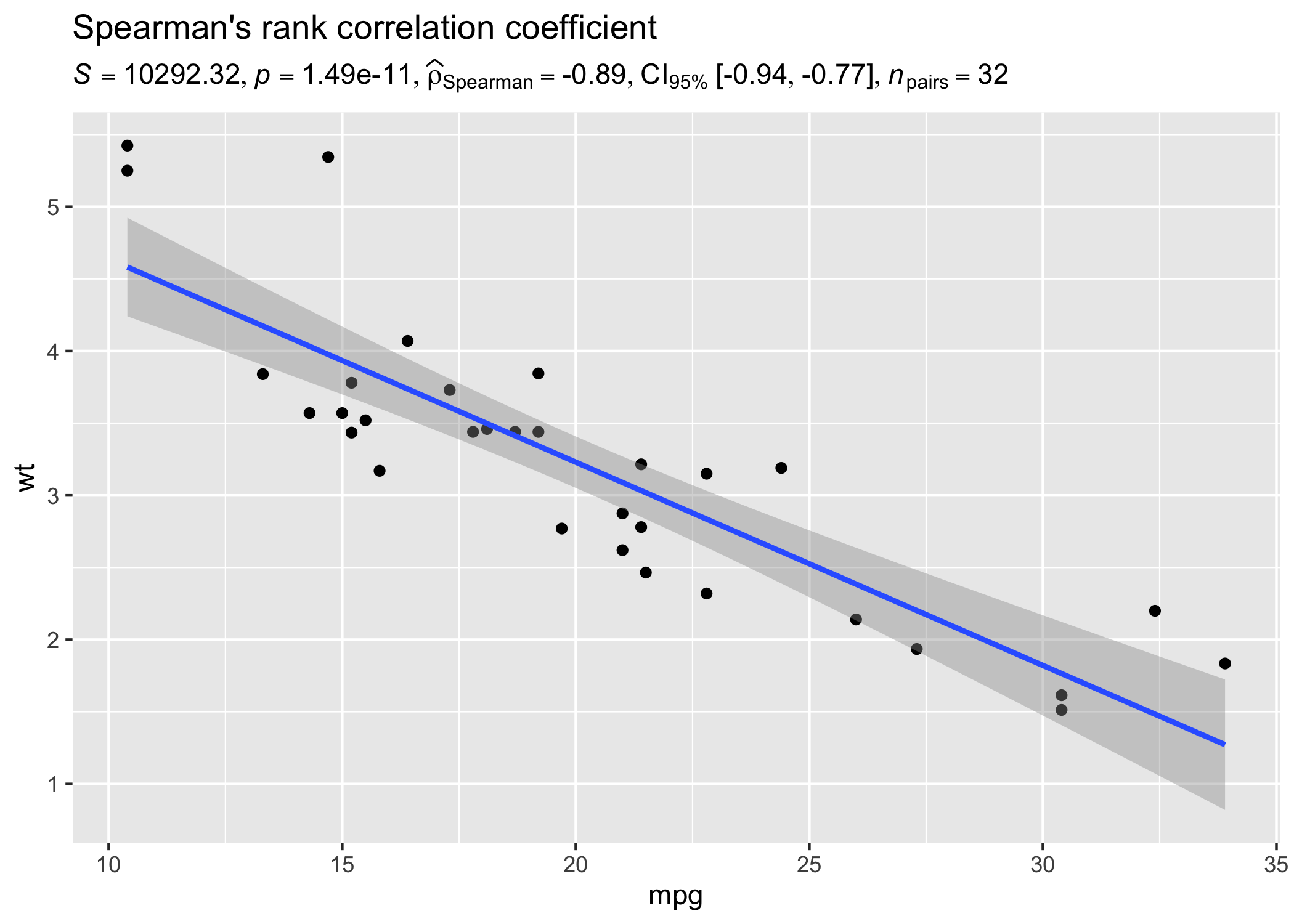
Let’s look at another example where we want to run correlation analysis:
set.seed(123)
# dataframe with results
results_data <- corr_test(mtcars, mpg, wt, type = "nonparametric")
# create a scatter plot
ggplot(mtcars, aes(mpg, wt)) +
geom_point() +
geom_smooth(method = "lm", formula = y ~ x) +
labs(
title = "Spearman's rank correlation coefficient",
subtitle = parse(text = results_data$expression)
)
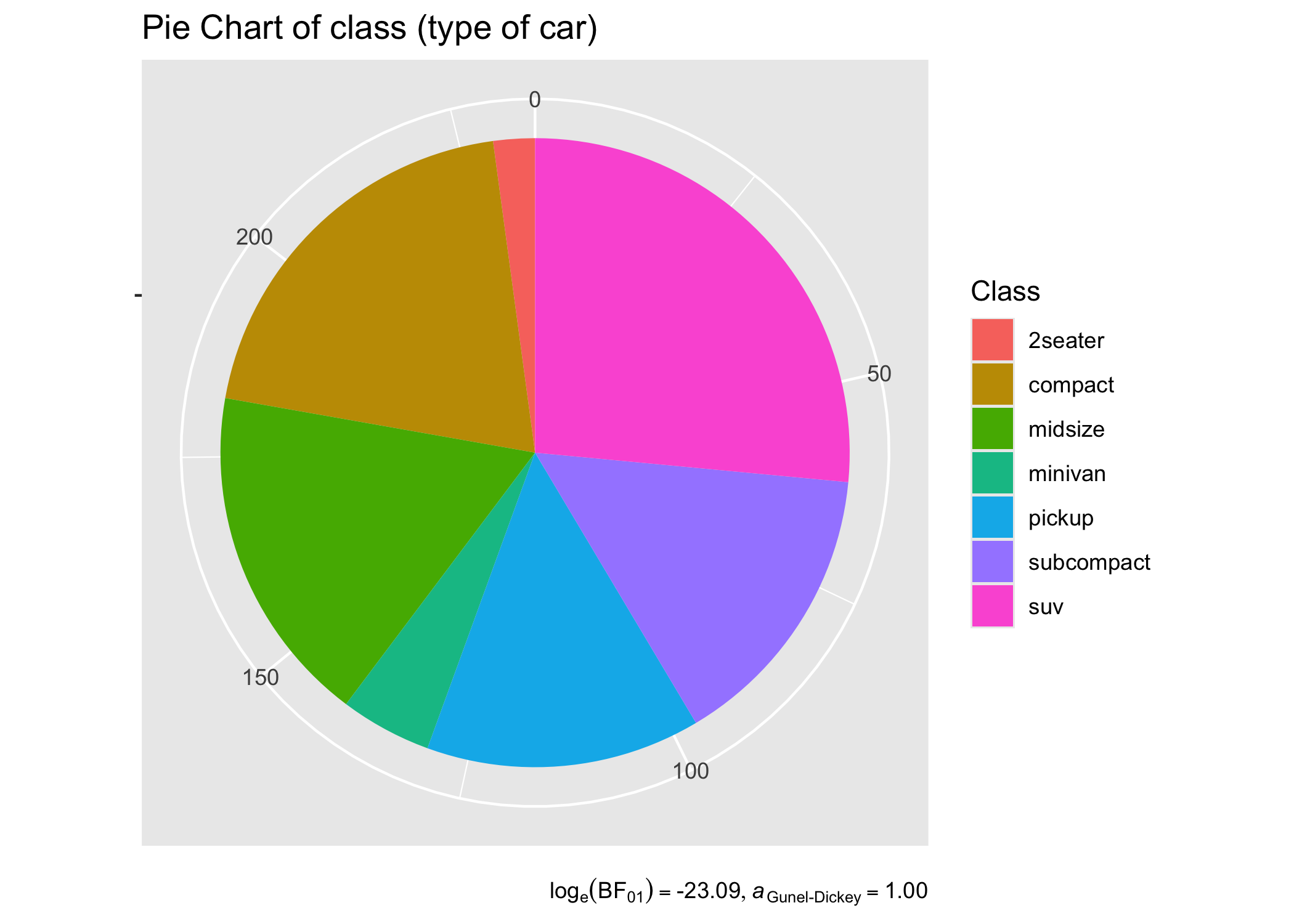
For categorical/nominal data - one-sample:
set.seed(123)
# dataframe with results
results_data <- contingency_table(
as.data.frame(table(mpg$class)),
Var1,
counts = Freq,
type = "bayes"
)
# create a pie chart
ggplot(as.data.frame(table(mpg$class)), aes(x = "", y = Freq, fill = factor(Var1))) +
geom_bar(width = 1, stat = "identity") +
theme(axis.line = element_blank()) +
# cleaning up the chart and adding results from one-sample proportion test
coord_polar(theta = "y", start = 0) +
labs(
fill = "Class",
x = NULL,
y = NULL,
title = "Pie Chart of class (type of car)",
caption = parse(text = results_data$expression)
)
You can also use these function to get the expression in return without having to display them in plots:
set.seed(123)
# Pearson's chi-squared test of independence
contingency_table(mtcars, am, vs)$expression[[1]]
#> list(chi["Pearson"]^2 * "(" * 1 * ")" == "0.91", italic(p) ==
#> "0.34", widehat(italic("V"))["Cramer"] == "0.00", CI["95%"] ~
#> "[" * "0.00", "0.49" * "]", italic("n")["obs"] == "32")set.seed(123)
library(metaviz)
# dataframe with results
results_data <- meta_analysis(dplyr::rename(mozart, estimate = d, std.error = se))
# meta-analysis forest plot with results random-effects meta-analysis
suppressWarnings(viz_forest(
x = mozart[, c("d", "se")],
study_labels = mozart[, "study_name"],
xlab = "Cohen's d",
variant = "thick",
type = "cumulative"
)) +
labs(
title = "Meta-analysis of Pietschnig, Voracek, and Formann (2010) on the Mozart effect",
subtitle = parse(text = results_data$expression)
) +
theme(text = element_text(size = 12))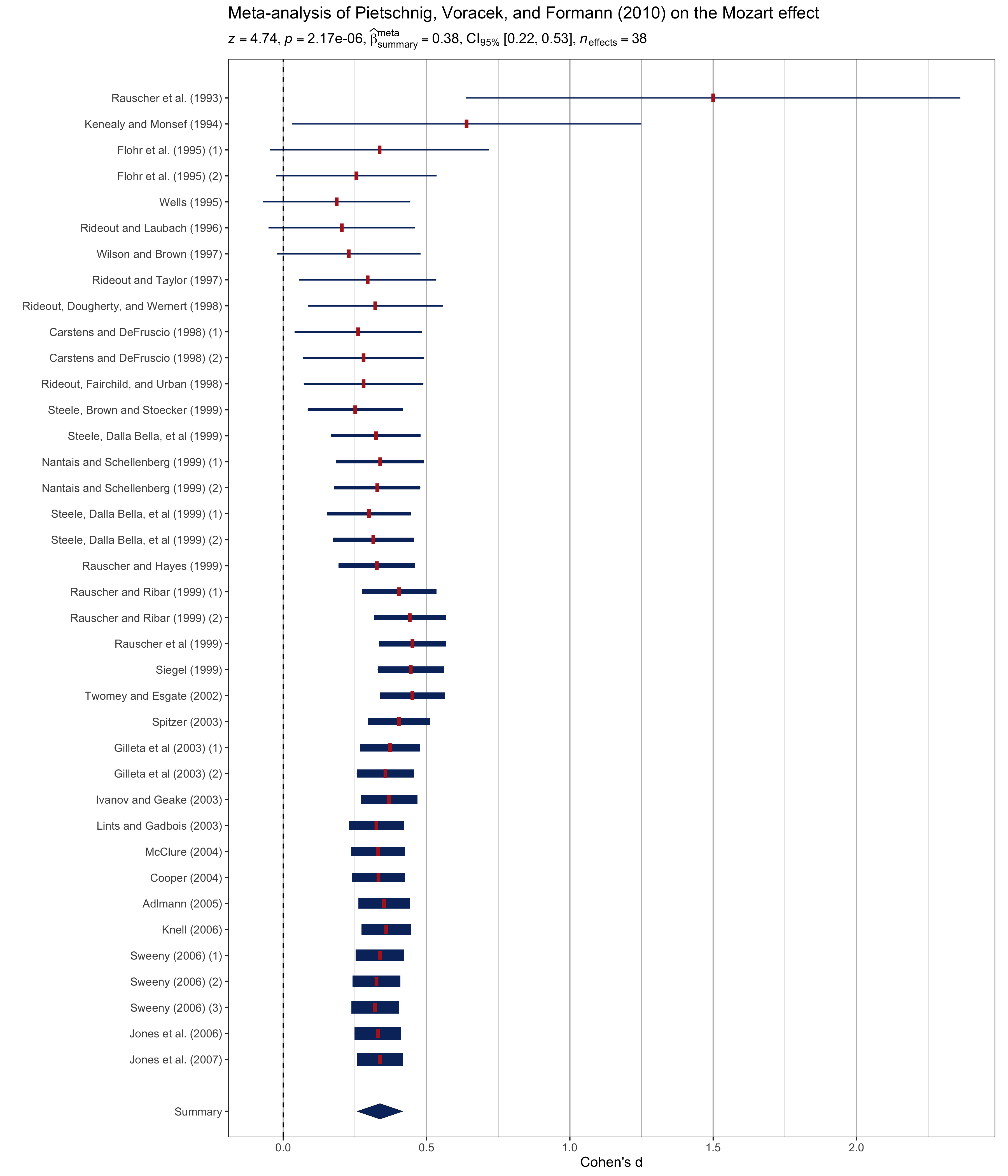
Sometimes you may not wish include so many details in the subtitle. In that case, you can extract the expression and copy-paste only the part you wish to include. For example, here only statistic and p-values are included:
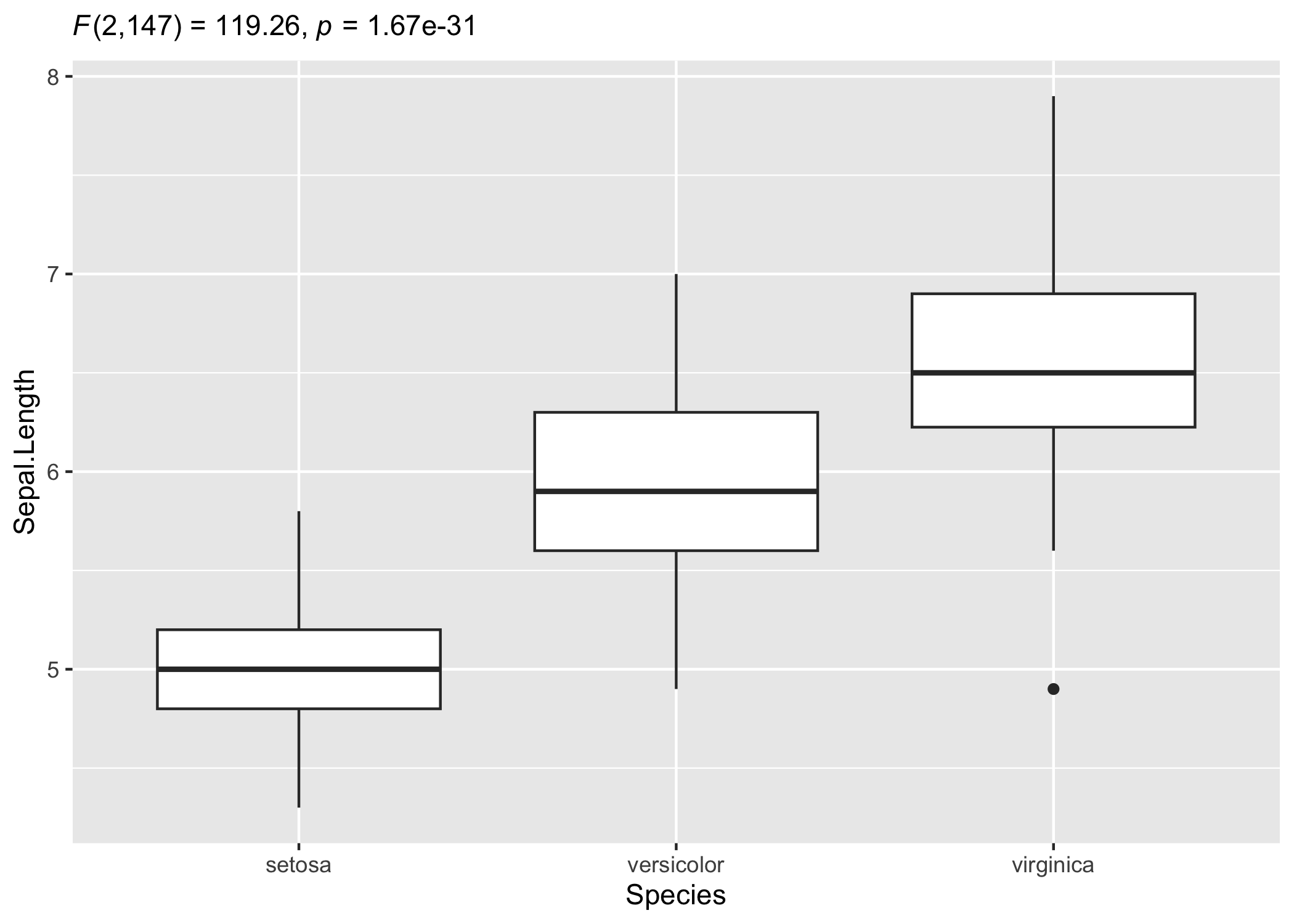
set.seed(123)
# extracting detailed expression
(res_expr <- oneway_anova(iris, Species, Sepal.Length, var.equal = TRUE)$expression[[1]])
#> list(italic("F")["Fisher"](2, 147) == "119.26", italic(p) ==
#> "1.67e-31", widehat(omega["p"]^2) == "0.61", CI["95%"] ~
#> "[" * "0.53", "1.00" * "]", italic("n")["obs"] == "150")
# adapting the details to your liking
ggplot(iris, aes(x = Species, y = Sepal.Length)) +
geom_boxplot() +
labs(subtitle = ggplot2::expr(paste(
NULL, italic("F"), "(", "2", ",", "147", ") = ", "119.26", ", ",
italic("p"), " = ", "1.67e-31"
)))
Here a go-to summary about statistical test carried out and the
returned effect size for each function is provided. This should be
useful if one needs to find out more information about how an argument
is resolved in the underlying package or if one wishes to browse the
source code. So, for example, if you want to know more about how one-way
(between-subjects) ANOVA, you can run ?stats::oneway.test
in your R console.
centrality_description| Type | Measure | Function used |
|---|---|---|
| Parametric | mean | datawizard::describe_distribution() |
| Non-parametric | median | datawizard::describe_distribution() |
| Robust | trimmed mean | datawizard::describe_distribution() |
| Bayesian | MAP | datawizard::describe_distribution() |
oneway_anovaHypothesis testing
| Type | No. of groups | Test | Function used |
|---|---|---|---|
| Parametric | > 2 | Fisher’s or Welch’s one-way ANOVA | stats::oneway.test() |
| Non-parametric | > 2 | Kruskal-Wallis one-way ANOVA | stats::kruskal.test() |
| Robust | > 2 | Heteroscedastic one-way ANOVA for trimmed means | WRS2::t1way() |
| Bayes Factor | > 2 | Fisher’s ANOVA | BayesFactor::anovaBF() |
Effect size estimation
| Type | No. of groups | Effect size | CI available? | Function used |
|---|---|---|---|---|
| Parametric | > 2 | partial eta-squared, partial omega-squared | Yes | effectsize::omega_squared(),
effectsize::eta_squared() |
| Non-parametric | > 2 | rank epsilon squared | Yes | effectsize::rank_epsilon_squared() |
| Robust | > 2 | Explanatory measure of effect size | Yes | WRS2::t1way() |
| Bayes Factor | > 2 | Bayesian R-squared | Yes | performance::r2_bayes() |
Hypothesis testing
| Type | No. of groups | Test | Function used |
|---|---|---|---|
| Parametric | > 2 | One-way repeated measures ANOVA | afex::aov_ez() |
| Non-parametric | > 2 | Friedman rank sum test | stats::friedman.test() |
| Robust | > 2 | Heteroscedastic one-way repeated measures ANOVA for trimmed means | WRS2::rmanova() |
| Bayes Factor | > 2 | One-way repeated measures ANOVA | BayesFactor::anovaBF() |
Effect size estimation
| Type | No. of groups | Effect size | CI available? | Function used |
|---|---|---|---|---|
| Parametric | > 2 | partial eta-squared, partial omega-squared | Yes | effectsize::omega_squared(),
effectsize::eta_squared() |
| Non-parametric | > 2 | Kendall’s coefficient of concordance | Yes | effectsize::kendalls_w() |
| Robust | > 2 | Algina-Keselman-Penfield robust standardized difference average | Yes | WRS2::wmcpAKP() |
| Bayes Factor | > 2 | Bayesian R-squared | Yes | performance::r2_bayes() |
two_sample_testHypothesis testing
| Type | No. of groups | Test | Function used |
|---|---|---|---|
| Parametric | 2 | Student’s or Welch’s t-test | stats::t.test() |
| Non-parametric | 2 | Mann-Whitney U test | stats::wilcox.test() |
| Robust | 2 | Yuen’s test for trimmed means | WRS2::yuen() |
| Bayesian | 2 | Student’s t-test | BayesFactor::ttestBF() |
Effect size estimation
| Type | No. of groups | Effect size | CI available? | Function used |
|---|---|---|---|---|
| Parametric | 2 | Cohen’s d, Hedge’s g | Yes | effectsize::cohens_d(),
effectsize::hedges_g() |
| Non-parametric | 2 | r (rank-biserial correlation) | Yes | effectsize::rank_biserial() |
| Robust | 2 | Algina-Keselman-Penfield robust standardized difference | Yes | WRS2::akp.effect() |
| Bayesian | 2 | difference | Yes | bayestestR::describe_posterior() |
Hypothesis testing
| Type | No. of groups | Test | Function used |
|---|---|---|---|
| Parametric | 2 | Student’s t-test | stats::t.test() |
| Non-parametric | 2 | Wilcoxon signed-rank test | stats::wilcox.test() |
| Robust | 2 | Yuen’s test on trimmed means for dependent samples | WRS2::yuend() |
| Bayesian | 2 | Student’s t-test | BayesFactor::ttestBF() |
Effect size estimation
| Type | No. of groups | Effect size | CI available? | Function used |
|---|---|---|---|---|
| Parametric | 2 | Cohen’s d, Hedge’s g | Yes | effectsize::cohens_d(),
effectsize::hedges_g() |
| Non-parametric | 2 | r (rank-biserial correlation) | Yes | effectsize::rank_biserial() |
| Robust | 2 | Algina-Keselman-Penfield robust standardized difference | Yes | WRS2::wmcpAKP() |
| Bayesian | 2 | difference | Yes | bayestestR::describe_posterior() |
one_sample_testHypothesis testing
| Type | Test | Function used |
|---|---|---|
| Parametric | One-sample Student’s t-test | stats::t.test() |
| Non-parametric | One-sample Wilcoxon test | stats::wilcox.test() |
| Robust | Bootstrap-t method for one-sample test | WRS2::trimcibt() |
| Bayesian | One-sample Student’s t-test | BayesFactor::ttestBF() |
Effect size estimation
| Type | Effect size | CI available? | Function used |
|---|---|---|---|
| Parametric | Cohen’s d, Hedge’s g | Yes | effectsize::cohens_d(),
effectsize::hedges_g() |
| Non-parametric | r (rank-biserial correlation) | Yes | effectsize::rank_biserial() |
| Robust | trimmed mean | Yes | WRS2::trimcibt() |
| Bayes Factor | difference | Yes | bayestestR::describe_posterior() |
corr_testHypothesis testing and Effect size estimation
| Type | Test | CI available? | Function used |
|---|---|---|---|
| Parametric | Pearson’s correlation coefficient | Yes | correlation::correlation() |
| Non-parametric | Spearman’s rank correlation coefficient | Yes | correlation::correlation() |
| Robust | Winsorized Pearson’s correlation coefficient | Yes | correlation::correlation() |
| Bayesian | Bayesian Pearson’s correlation coefficient | Yes | correlation::correlation() |
contingency_tableHypothesis testing
| Type | Design | Test | Function used |
|---|---|---|---|
| Parametric/Non-parametric | Unpaired | Pearson’s chi-squared test | stats::chisq.test() |
| Bayesian | Unpaired | Bayesian Pearson’s chi-squared test | BayesFactor::contingencyTableBF() |
| Parametric/Non-parametric | Paired | McNemar’s chi-squared test | stats::mcnemar.test() |
| Bayesian | Paired | No | No |
Effect size estimation
| Type | Design | Effect size | CI available? | Function used |
|---|---|---|---|---|
| Parametric/Non-parametric | Unpaired | Cramer’s V | Yes | effectsize::cramers_v() |
| Bayesian | Unpaired | Cramer’s V | Yes | effectsize::cramers_v() |
| Parametric/Non-parametric | Paired | Cohen’s g | Yes | effectsize::cohens_g() |
| Bayesian | Paired | No | No | No |
Hypothesis testing
| Type | Test | Function used |
|---|---|---|
| Parametric/Non-parametric | Goodness of fit chi-squared test | stats::chisq.test() |
| Bayesian | Bayesian Goodness of fit chi-squared test | (custom) |
Effect size estimation
| Type | Effect size | CI available? | Function used |
|---|---|---|---|
| Parametric/Non-parametric | Pearson’s C | Yes | effectsize::pearsons_c() |
| Bayesian | No | No | No |
meta_analysisHypothesis testing and Effect size estimation
| Type | Test | Effect size | CI available? | Function used |
|---|---|---|---|---|
| Parametric | Meta-analysis via random-effects models | beta | Yes | metafor::metafor() |
| Robust | Meta-analysis via robust random-effects models | beta | Yes | metaplus::metaplus() |
| Bayes | Meta-analysis via Bayesian random-effects models | beta | Yes | metaBMA::meta_random() |
{ggstatsplot}Note that these functions were initially written to display results
from statistical tests on ready-made {ggplot2} plots
implemented in {ggstatsplot}.
For detailed documentation, see the package website: https://indrajeetpatil.github.io/ggstatsplot/
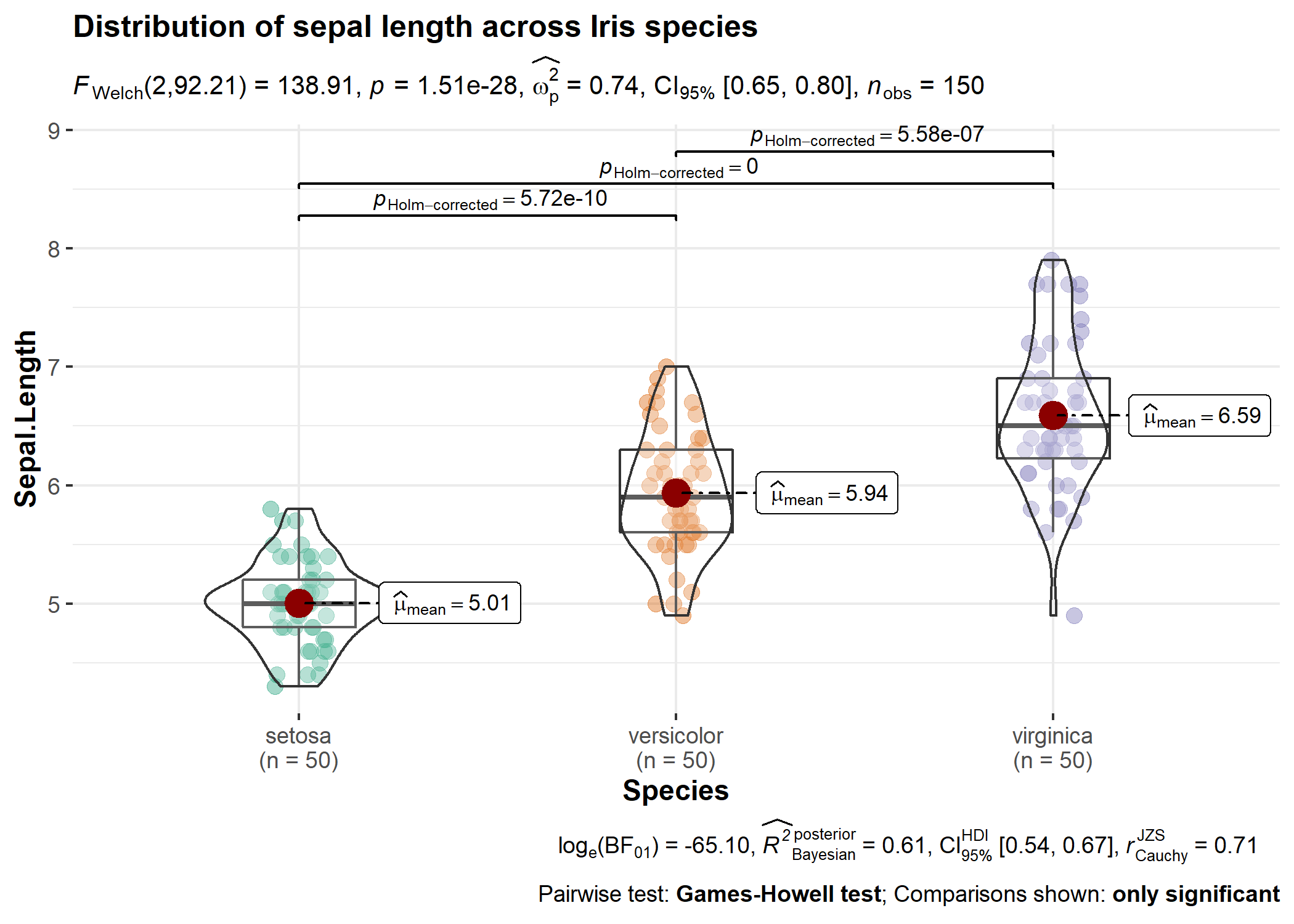
Here is an example from {ggstatsplot} of what the plots
look like when the expressions are displayed in the subtitle-
The hexsticker and the schematic illustration of general workflow were generously designed by Sarah Otterstetter (Max Planck Institute for Human Development, Berlin).
Bug reports, suggestions, questions, and (most of all) contributions are welcome.
Please note that this project is released with a Contributor Code of Conduct. By participating in this project you agree to abide by its terms.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.