The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.
The goal of tidyplots is to streamline the creation of
publication-ready plots for scientific papers. It allows to gradually
add, remove and adjust plot components using a consistent and intuitive
syntax.
Engler, Jan Broder. 2025. “Tidyplots Empowers Life Scientists With Easy Code-Based Data Visualization.” iMeta e70018. https://doi.org/10.1002/imt2.70018
You can install the released version of tidyplots from CRAN with:
install.packages("tidyplots")And the development version from GitHub with:
# install.packages("pak")
pak::pak("jbengler/tidyplots")This cheatsheet gives a high level overview of available functions.
Here are some examples.
Also have a look at the getting started guide and the full documentation. For more example plots, check out the tidyplots use cases website.
library(tidyplots)
study |>
tidyplot(x = treatment, y = score, color = treatment) |>
add_mean_bar(alpha = 0.4) |>
add_sem_errorbar() |>
add_data_points_beeswarm()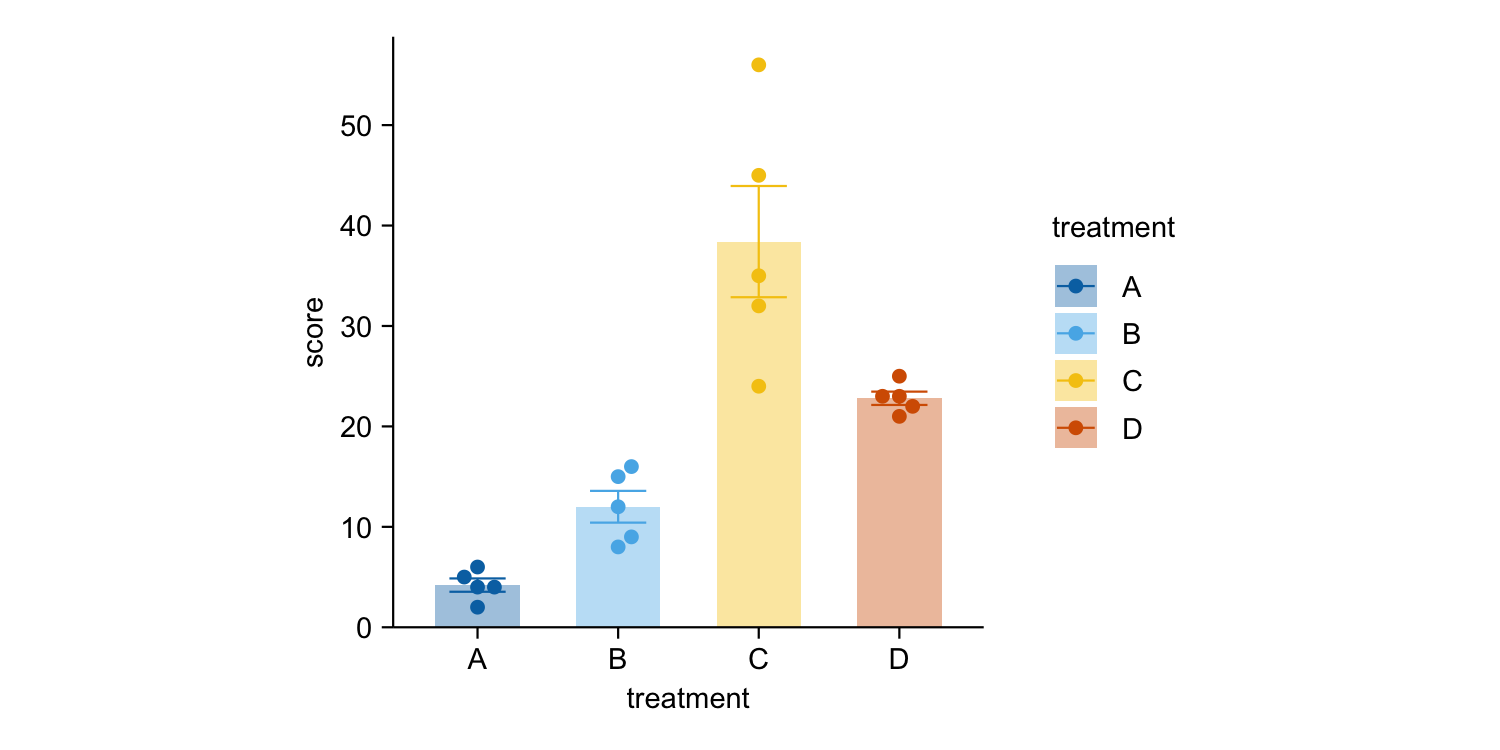
energy |>
tidyplot(x = year, y = energy, color = energy_source) |>
add_barstack_absolute()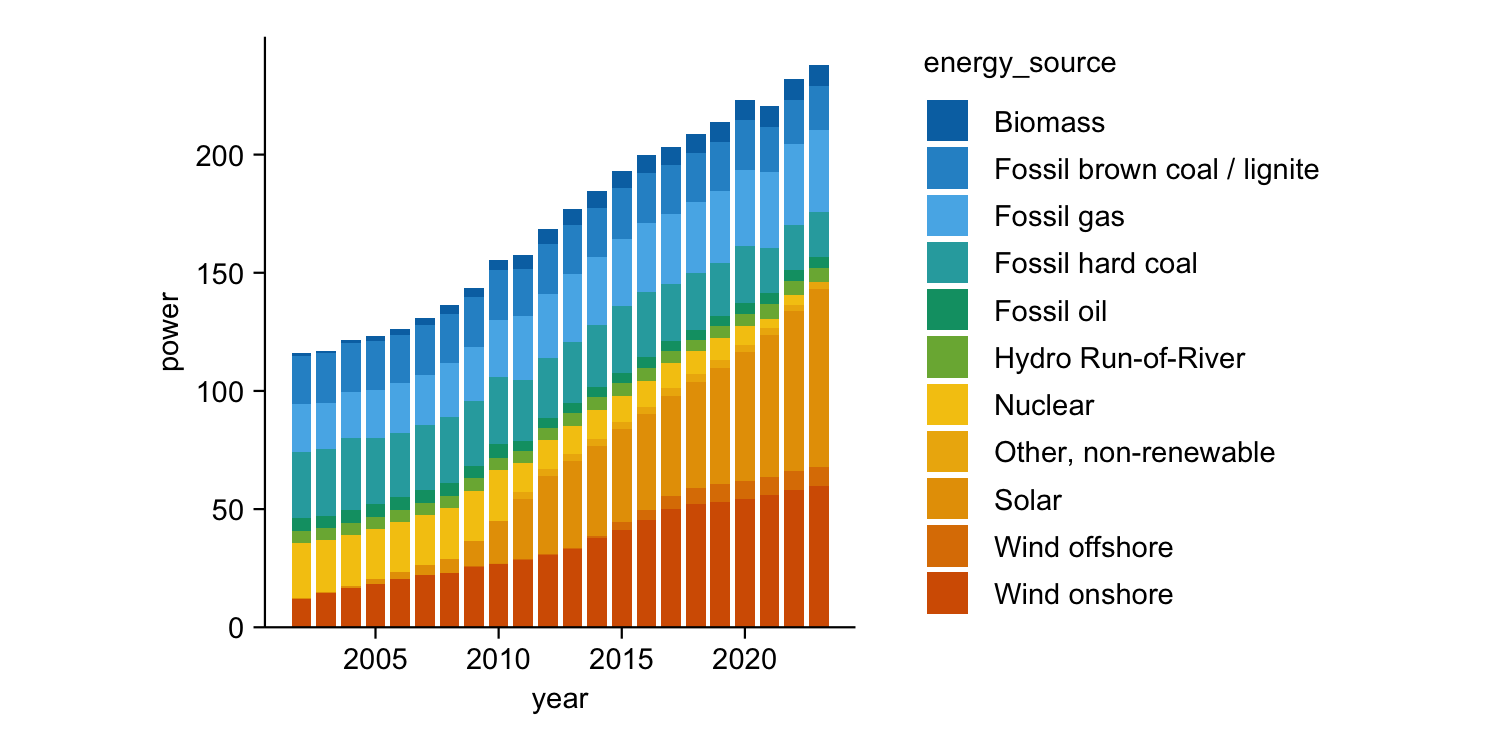
energy |>
dplyr::filter(year %in% c(2005, 2010, 2015, 2020)) |>
tidyplot(y = energy, color = energy_source) |>
add_donut() |>
adjust_size(width = 25, height = 25) |>
split_plot(by = year)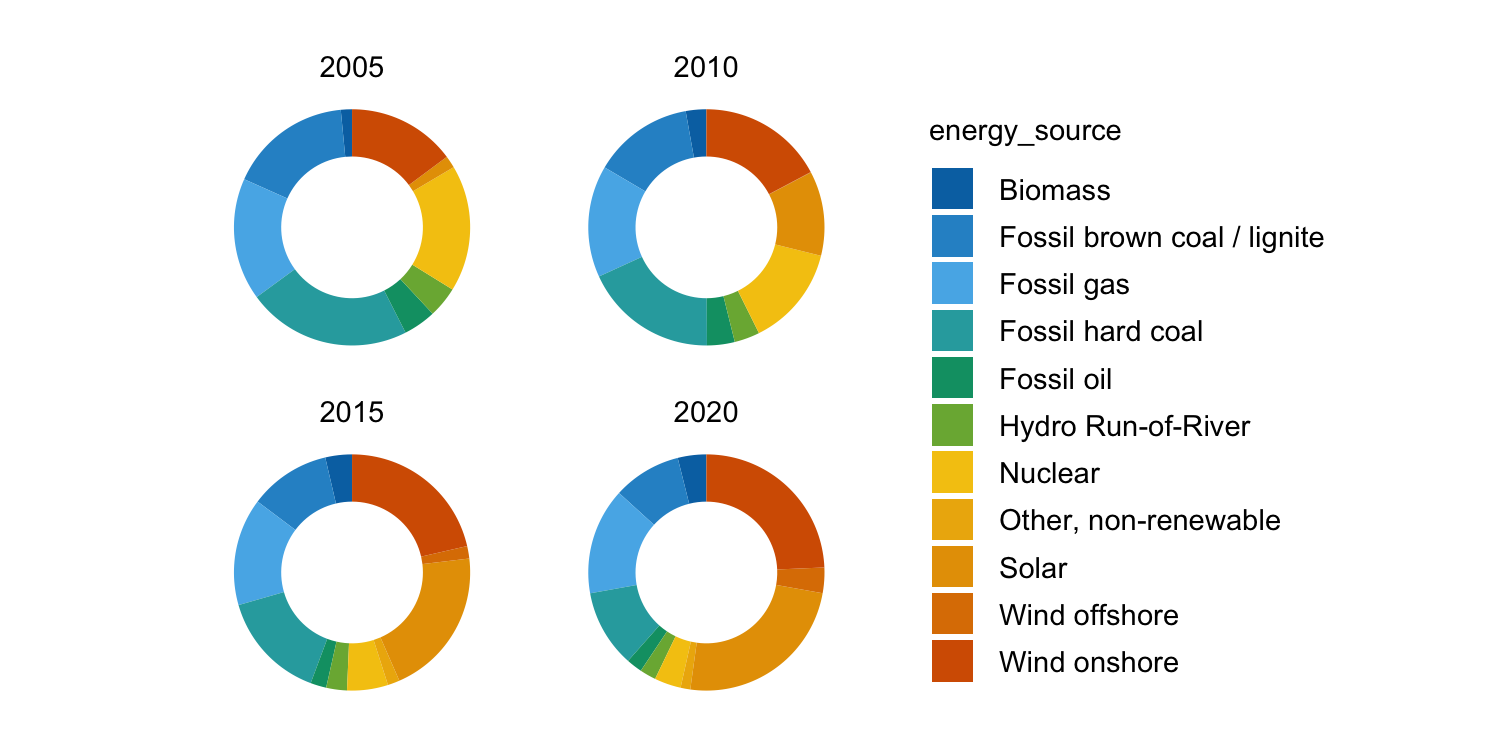
energy_week |>
tidyplot(x = date, y = power, color = energy_source) |>
add_areastack_absolute()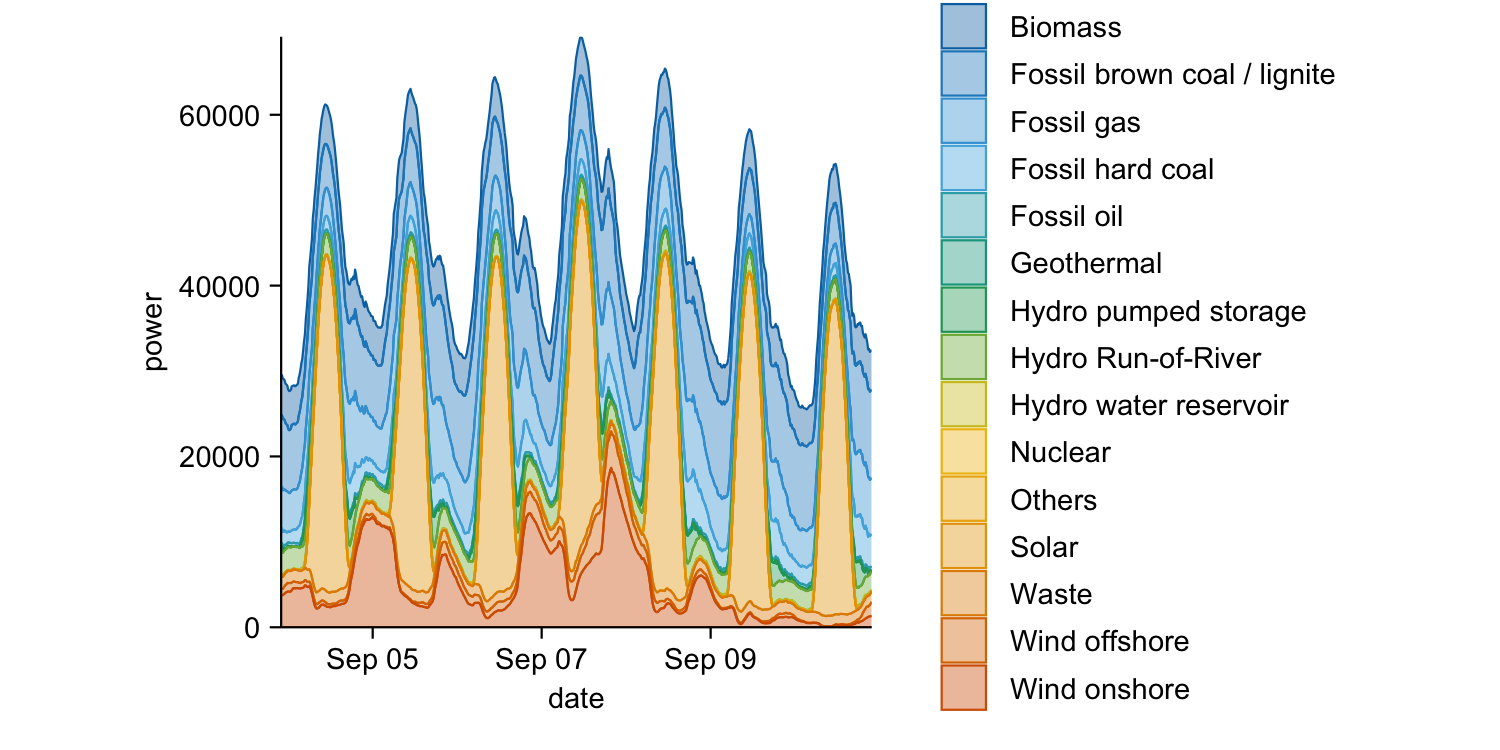
energy_week |>
tidyplot(x = date, y = power, color = energy_source) |>
add_areastack_relative()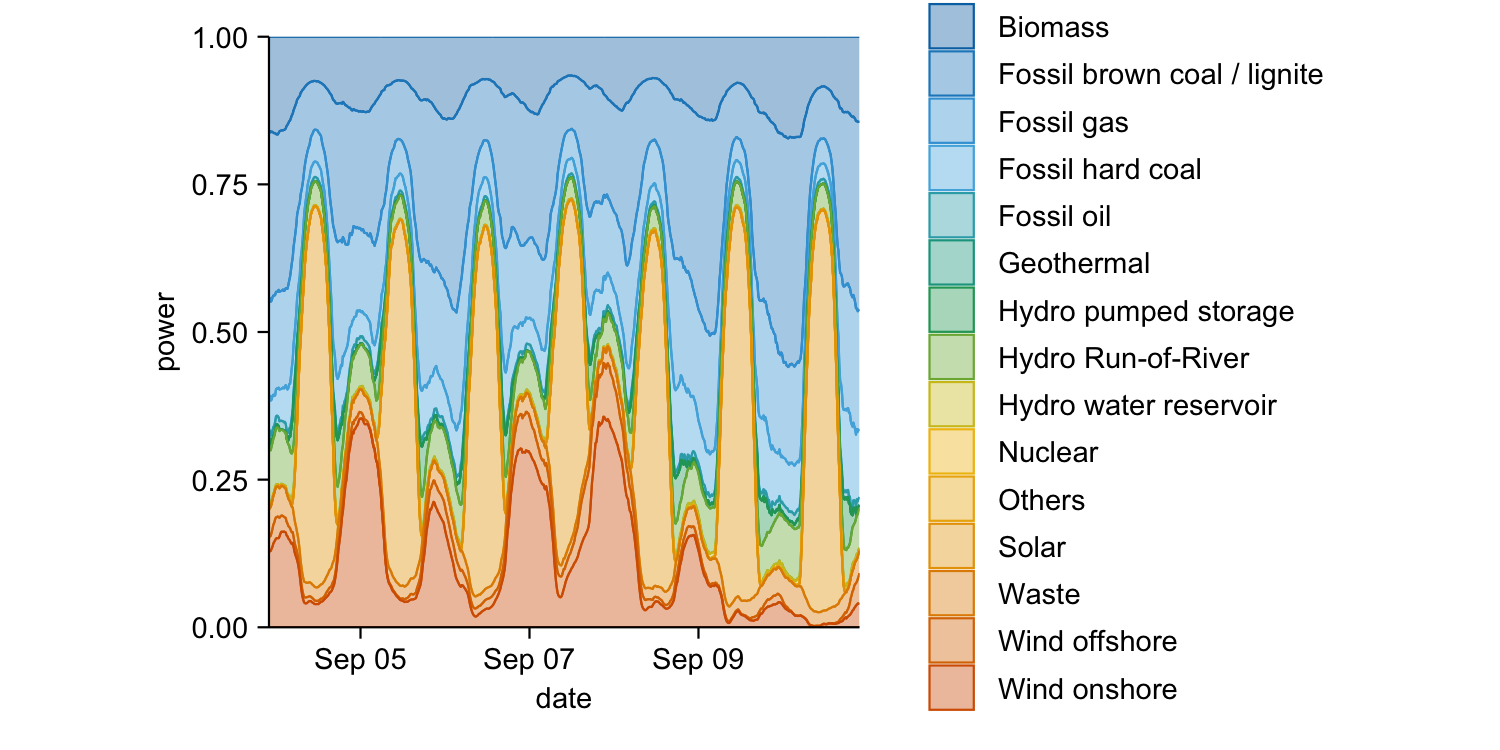
study |>
tidyplot(x = group, y = score, color = dose) |>
add_mean_bar(alpha = 0.4) |>
add_mean_dash() |>
add_mean_value()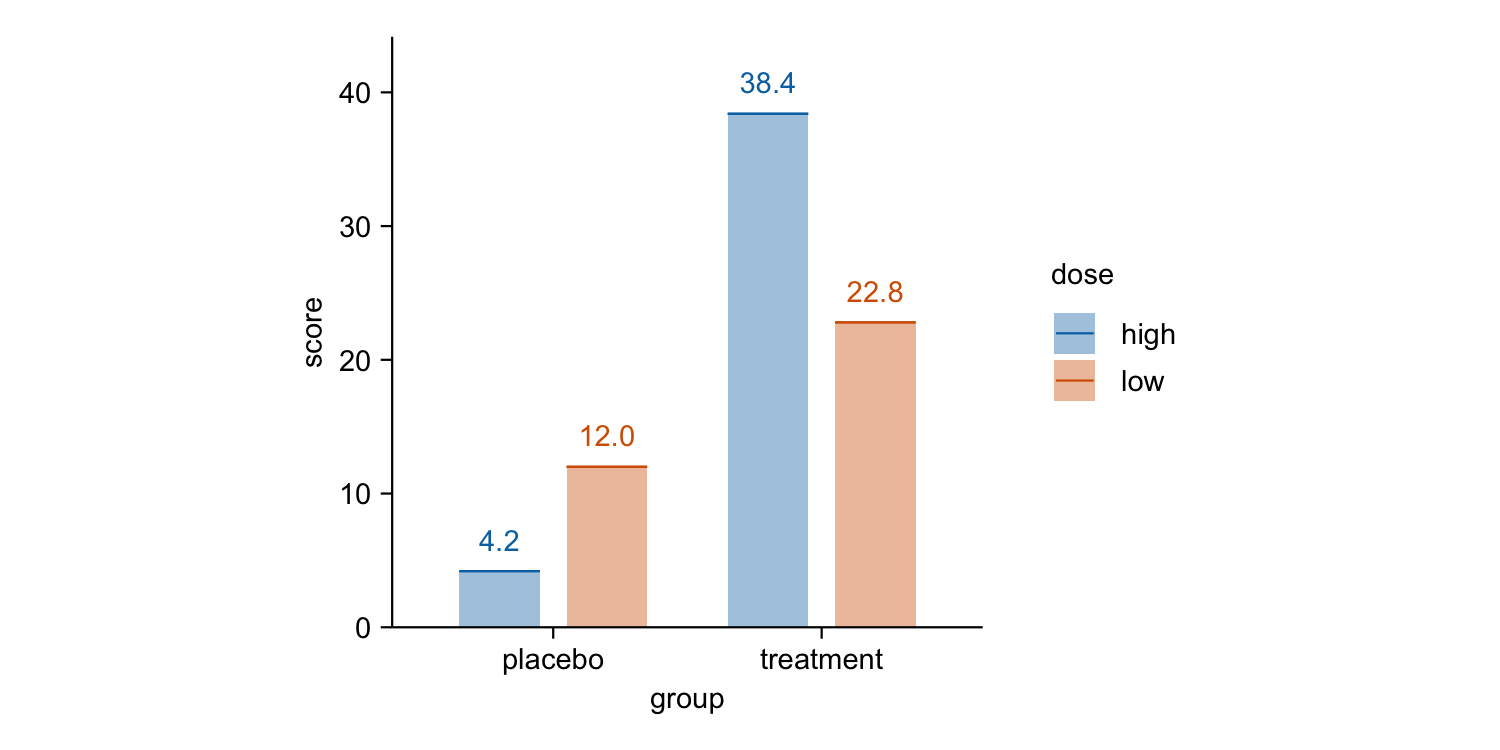
time_course |>
tidyplot(x = day, y = score, color = treatment) |>
add_mean_line() |>
add_mean_dot() |>
add_sem_ribbon()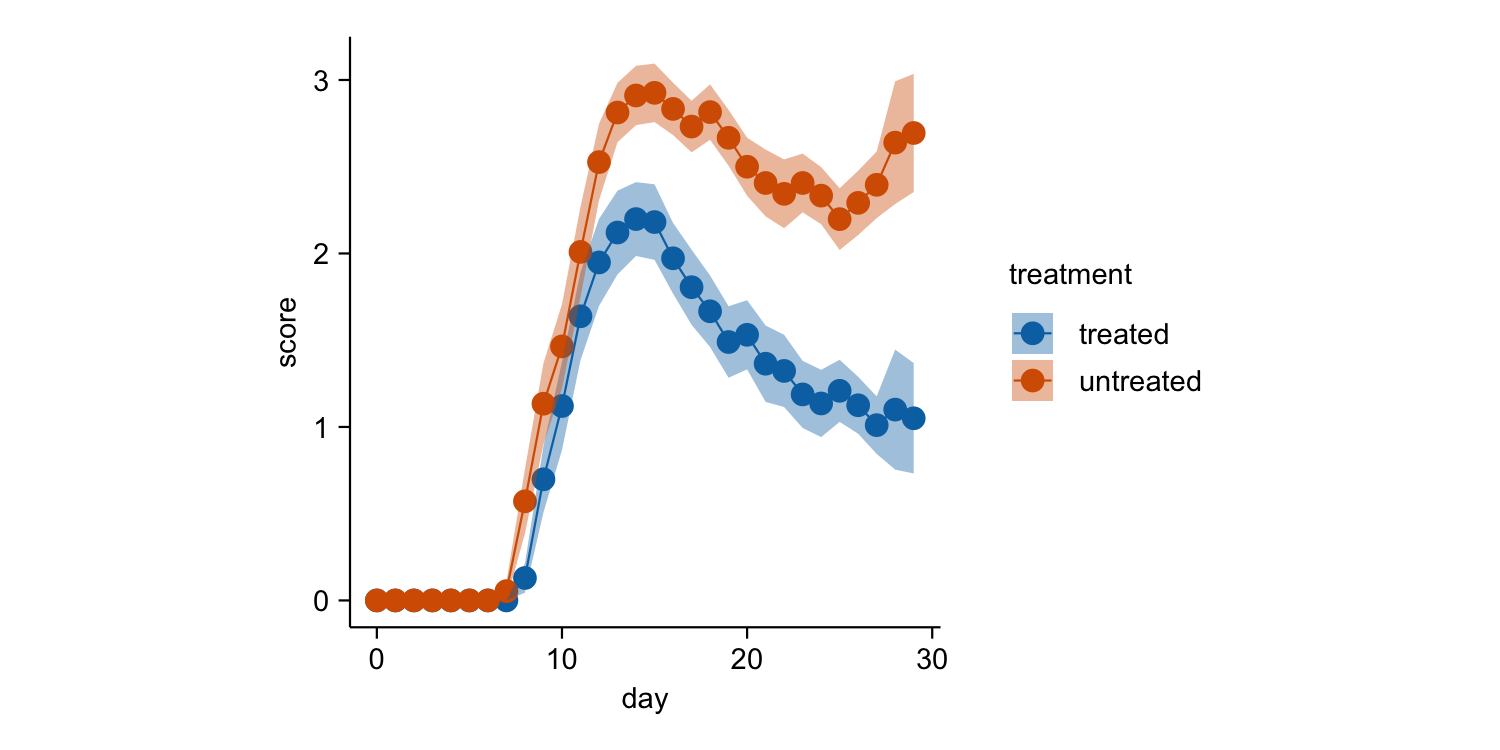
climate |>
tidyplot(x = month, y = year, color = max_temperature) |>
add_heatmap()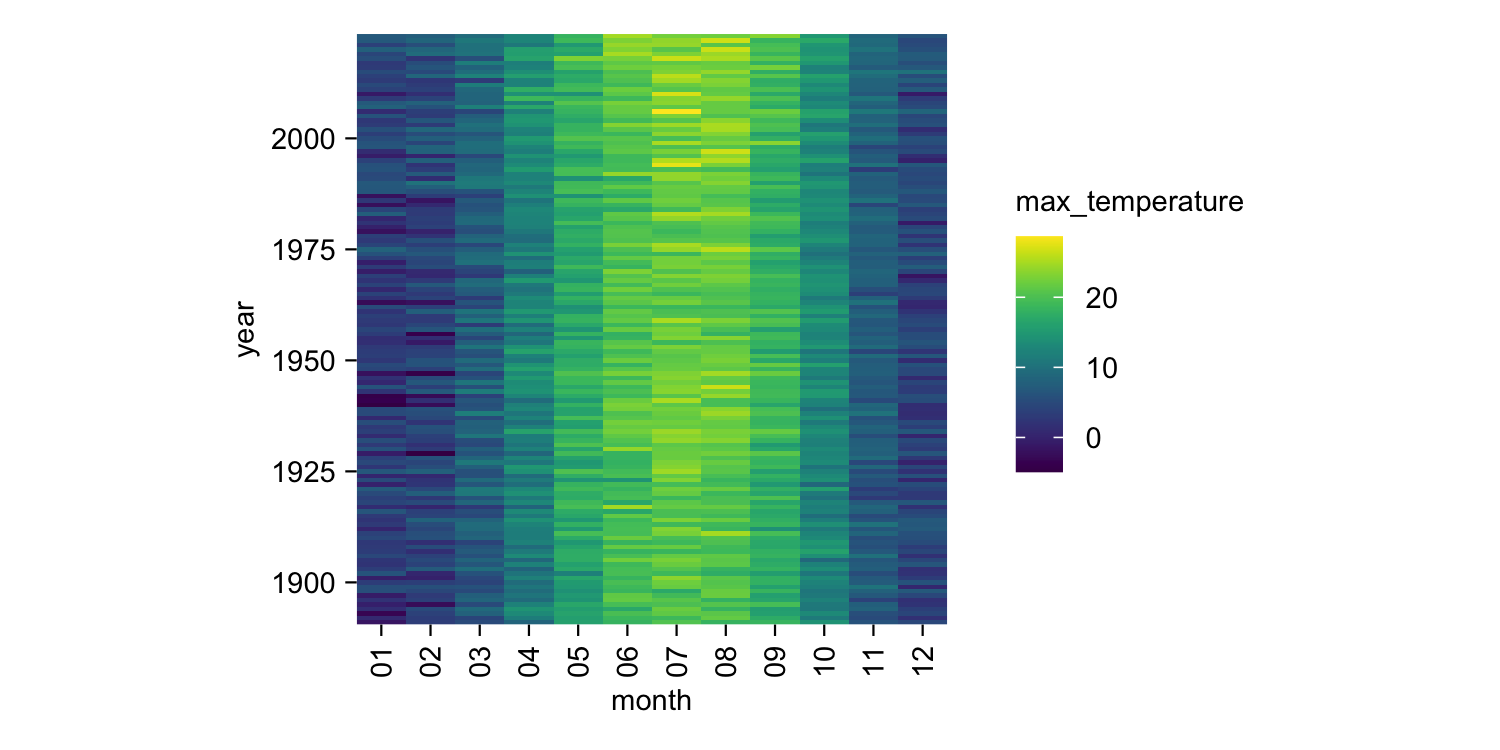
study |>
tidyplot(x = treatment, y = score, color = treatment) |>
add_boxplot() |>
add_test_pvalue(ref.group = 1)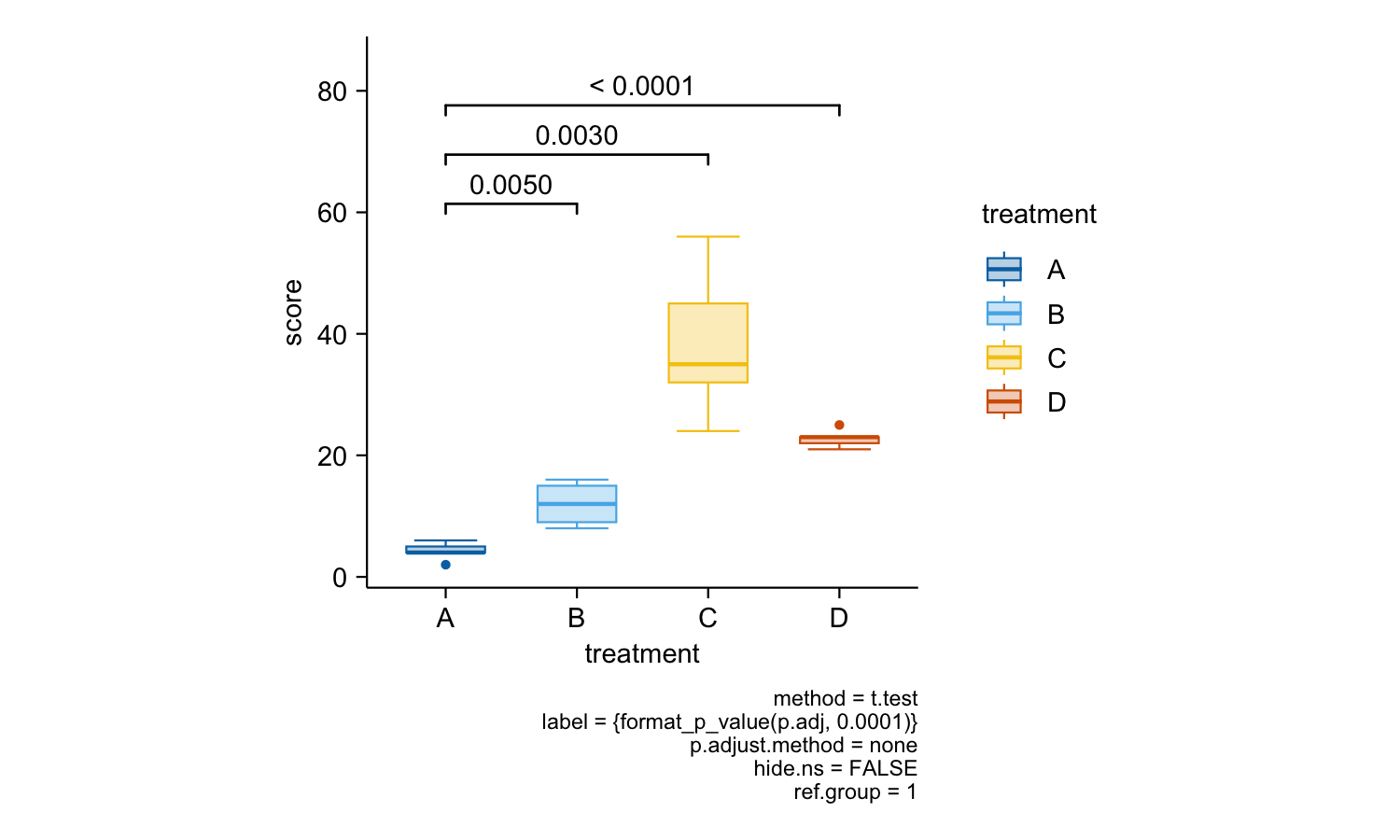
gene_expression |>
dplyr::filter(external_gene_name %in% c("Apol6", "Col5a3", "Vgf", "Bsn")) |>
tidyplot(x = condition, y = expression, color = sample_type) |>
add_mean_dash() |>
add_sem_errorbar() |>
add_data_points_beeswarm() |>
add_test_asterisks(hide_info = TRUE) |>
remove_x_axis_title() |>
adjust_size(width = 25, height = 25) |>
split_plot(by = external_gene_name)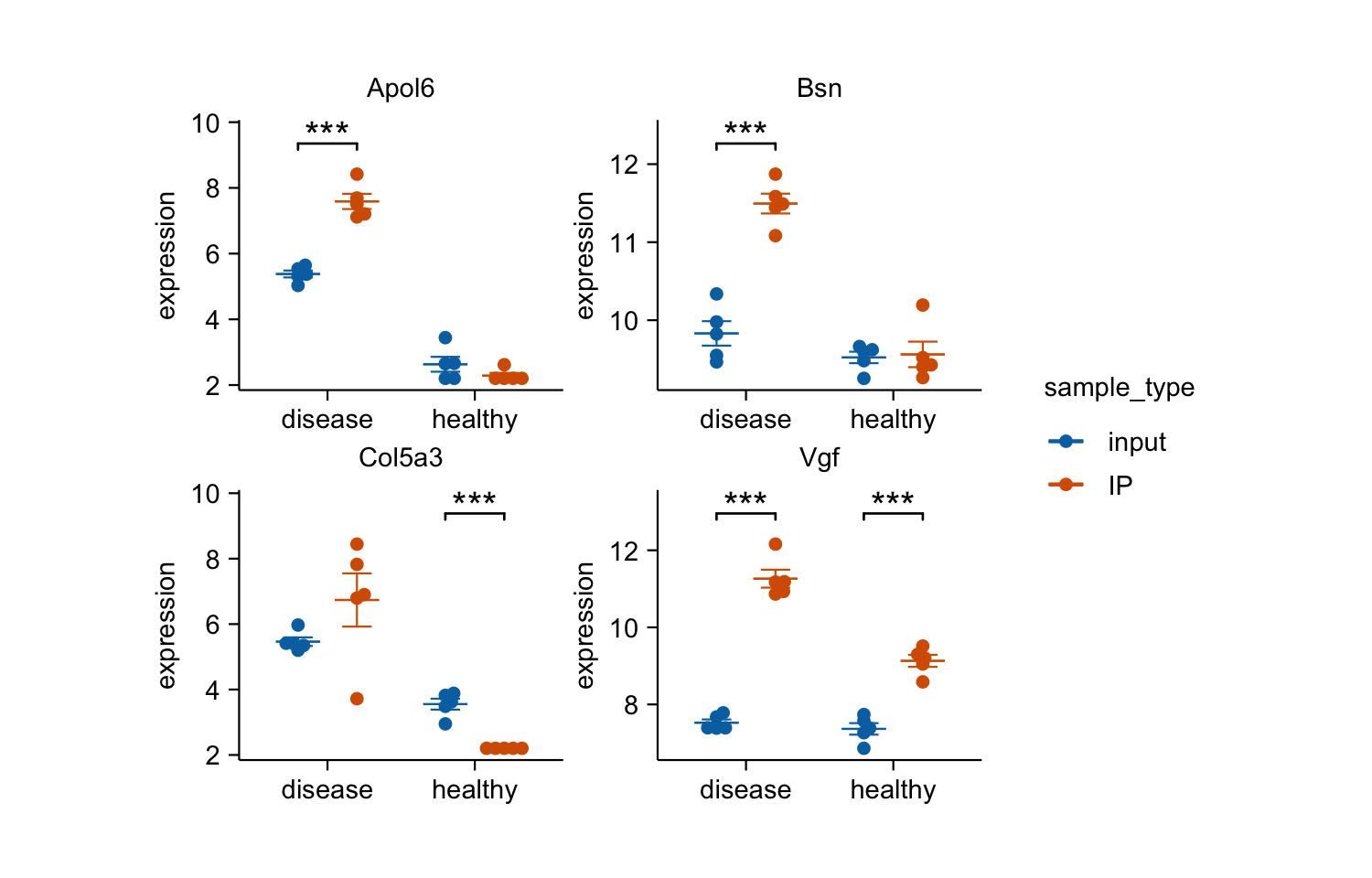
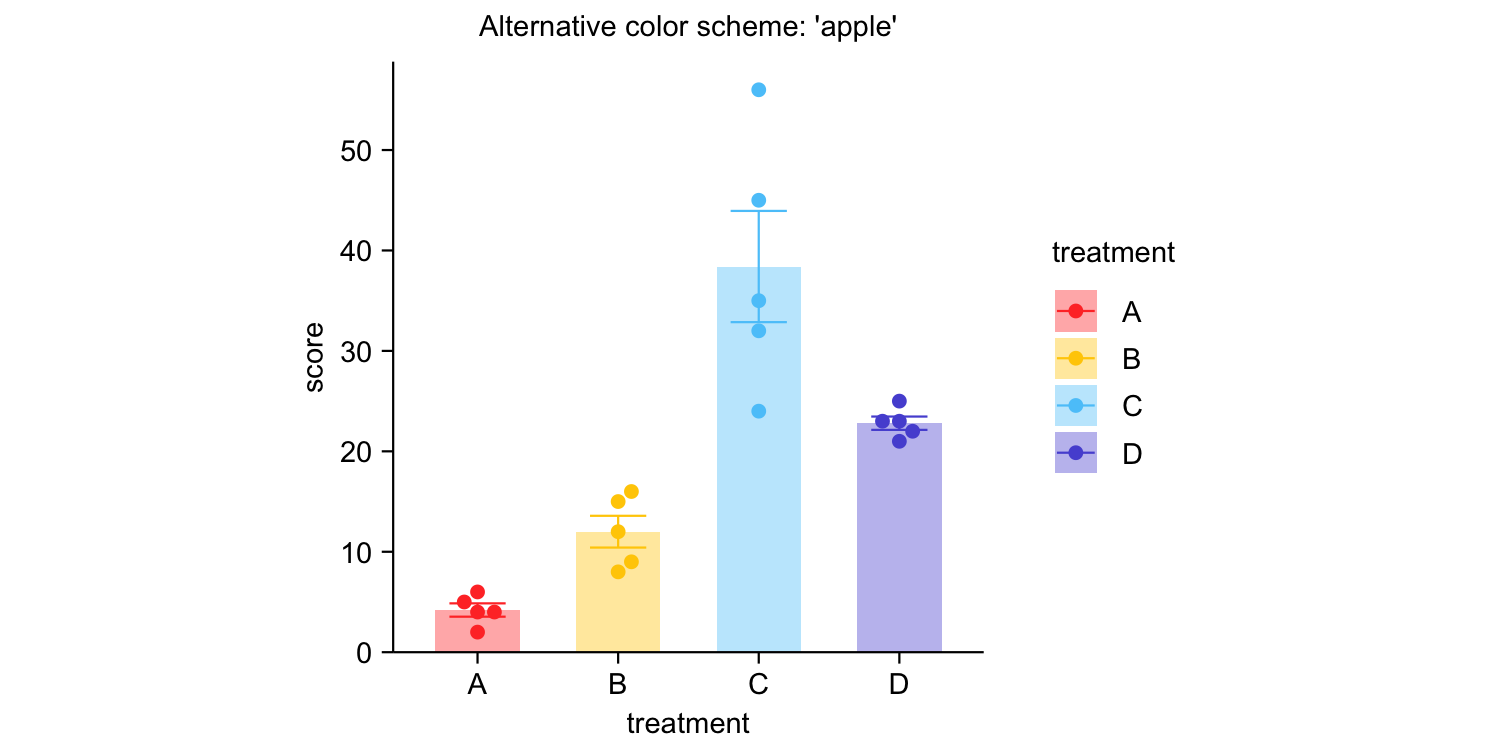
study |>
tidyplot(x = treatment, y = score, color = treatment) |>
add_mean_bar(alpha = 0.4) |>
add_sem_errorbar() |>
add_data_points_beeswarm() |>
view_plot(title = "Default color scheme: 'friendly'") |>
adjust_colors(colors_discrete_apple) |>
view_plot(title = "Alternative color scheme: 'apple'")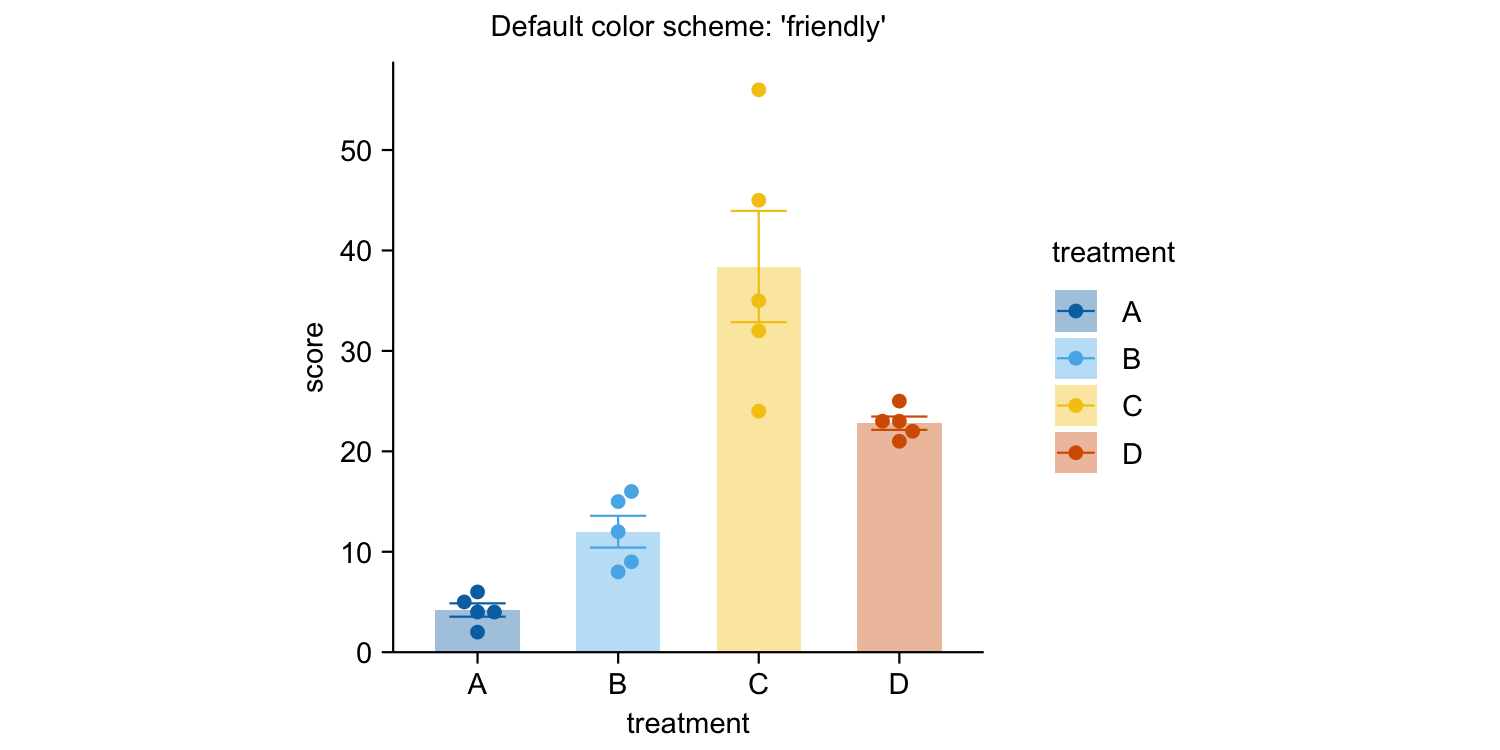
Package
index
Overview of all tidyplots functions
Get
started
Getting started guide
Visualizing
data
Article with examples for common data visualizations
Advanced
plotting
Article about advanced plotting techniques and workflows
Color
schemes
Article about the use of color schemes
I would like to thank Lars Binkle-Ladisch for our insightful discussions and for consistently challenging my decisions regarding the naming of functions and their arguments.
Many thanks to the R and tidyverse communities. tidyplots is built upon their software and coding paradigms, and it would not have been possible without their contributions.
tidyplots relies on several fantastic packages that handle all the heavy lifting behind the scenes. These include cli, dplyr, forcats, ggbeeswarm, ggplot2, ggpubr, ggrastr, ggrepel, glue, Hmisc, htmltools, lifecycle, purrr, rlang, scales, stringr, tidyr, and tidyselect.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.