The hardware and bandwidth for this mirror is donated by METANET, the Webhosting and Full Service-Cloud Provider.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]metanet.ch.

zscorer facilitates the calculation of a range of
anthropometric z-scores (i.e. the number of standard
deviations from the mean) and adds them to survey data:
Weight-for-length (wfl) z-scores for children with lengths between 45 and 110 cm
Weight-for-height (wfh) z-scores for children with heights between 65 and 120 cm
Length-for-age (lfa) z-scores for children aged less than 24 months
Height-for-age (hfa) z-scores for children aged between 24 and 228 months
Weight-for-age (wfa) z-scores for children aged between zero and 120 months
Body mass index-for-age (bfa) z-scores for children aged between zero and 228 months
MUAC-for-age (mfa) z-scores for children aged between 3 and 228 months
Triceps skinfold-for-age (tsa) z-scores for children aged between 3 and 60 months
Sub-scapular skinfold-for-age (ssa) z-scores for children aged between 3 and 60 months
Head circumference-for-age (hca) z-scores for children aged between zero and 60 months
The z-scores are calculated using the WHO Child
Growth Standards [1],[2] for children aged between zero and 60
months or the WHO Growth References [3] for school-aged
children and adolescents. MUAC-for-age (mfa) z-scores for children aged
between 60 and 228 months are calculated using the MUAC-for-age growth
reference developed by Mramba et al. (2017) [4] using data from the USA
and Africa. This reference has been validated with African school-age
children and adolescents. The zscorer comes packaged with
the WHO Growth References data and the MUAC-for-age reference data.
You can install zscorer from CRAN:
install.packages("zscorer")or you can install the development version of zscorer
from GitHub
with:
if(!require(remotes)) install.packages("remotes")
remotes::install_github("nutriverse/zscorer")then load zscorer
# load package
library(zscorer)The main function in the zscorer package is
addWGSR().
To demonstrate its usage, we will use the accompanying dataset in
zscorer called anthro3. We inspect the dataset
as follows:
head(anthro3)which returns:
#> psu age sex weight height muac oedema
#> 1 1 10 1 5.7 64.2 125 2
#> 2 1 10 2 5.8 64.4 121 2
#> 3 1 9 2 6.5 62.2 139 2
#> 4 1 11 9 6.5 64.9 129 2
#> 5 1 24 2 6.5 72.9 120 2
#> 6 1 12 2 6.6 69.4 126 2anthro3 contains anthropometric data from a Rapid
Assessment Method (RAM) survey from Burundi.
Anthropometric indices (e.g. weight-for-height z-scores) have not been calculated and added to the data.
We will use the addWGSR() function to add
weight-for-height (wfh) z-scores to the example data:
svy <- addWGSR(data = anthro3, sex = "sex", firstPart = "weight",
secondPart = "height", index = "wfh")
#> ===========================================================================A new column named wfhz has been added to the dataset:
#> psu age sex weight height muac oedema wfhz
#> 1 1 10 1 5.7 64.2 125 2 -2.73
#> 2 1 10 2 5.8 64.4 121 2 -2.04
#> 3 1 9 2 6.5 62.2 139 2 0.13
#> 4 1 11 9 6.5 64.9 129 2 NA
#> 5 1 24 2 6.5 72.9 120 2 -3.44
#> 6 1 12 2 6.6 69.4 126 2 -2.26The wfhz column contains the weight-for-height (wfh)
z-scores calculated from the sex, weight, and
height columns in the anthro3 dataset. The
calculated z-scores are rounded to two decimals places unless the
digits option is used to specify a different precision (run
?addWGSR to see description of various parameters that can
be specified in the addWGSR() function).
The addWGSR() function takes up to nine parameters to
calculate each index separately, depending on the index required. These
are described in the Help files of the zscorer
package which can be accessed as follows:
?addWGSRThe standing parameter specifies how “stature” (i.e. length or height) was measured. If this is not specified, and in some special circumstances, height and age rules will be applied when calculating z-scores. These rules are described in the table below.
| index | standing | age | height | Action |
|---|---|---|---|---|
| hfa or lfa | standing | < 731 days | index = lfa height = height + 0.7 cm | |
| hfa or lfa | supine | < 731 days | index = lfa | |
| hfa or lfa | unknown | < 731 days | index = lfa | |
| hfa or lfa | standing | ≥ 731 days | index = hfa | |
| hfa or lfa | supine | ≥ 731 days | index = hfa height = height - 0.7 cm | |
| hfa or lfa | unknown | ≥ 731 days | index = hfa | |
| wfh or wfl | standing | < 65 cm | index = wfl height = height + 0.7 cm | |
| wfh or wfl | standing | ≥ 65 cm | index = wfh | |
| wfh or wfl | supine | ≤ 110 cm | index = wfl | |
| wfh or wfl | supine | more than 110 cm | index = wfh height = height - 0.7 cm | |
| wfh or wfl | unknown | < 87 cm | index = wfl | |
| wfh or wfl | unknown | ≥ 87 cm | index = wfh | |
| bfa | standing | < 731 days | height = height + 0.7 cm | |
| bfa | standing | ≥ 731 days | height = height - 0.7 cm |
The addWGSR() function will not produce error messages
unless there is something very wrong with the data or the specified
parameters. If an error is encountered in a record then the value
NA is returned. Error conditions are listed in the
table below.
| Error condition | Action |
|---|---|
Missing or nonsense value in standing parameter |
Set standing to 3 (unknown) and apply
appropriate height or age rules. |
Unknown index specified |
Return NA for z-score. |
Missing sex |
Return NA for z-score. |
Missing firstPart |
Return NA for z-score. |
Missing secondPart |
Return NA for z-score. |
sex is not male (1) or female
(2) |
Return NA for z-score. |
firstPart is not numeric |
Return NA for z-score. |
secondPart is not numeric |
Return NA for z-score. |
Missing thirdPart when index = "bfa" |
Return NA for z-score. |
thirdPart is not numeric when
index = "bfa" |
Return NA for z-score. |
secondPart is out of range for specified index |
Return NA for z-score. |
We can see this error behaviour using the example data:
table(is.na(svy$wfhz))
#>
#> FALSE TRUE
#> 220 1We can display the problem record:
svy[is.na(svy$wfhz), ]
#> psu age sex weight height muac oedema wfhz
#> 4 1 11 9 6.5 64.9 129 2 NAThe problem is due to the value 9 in the
sex column, which should be coded 1 (for
male) and 2 (for female). Z-scores are only calculated
for records with sex specified as either 1 (male) or
2 (female). All other values, including
NA, will return NA.
The addWGSR() function requires that data are recorded
using the required units or required codes (see ?addWGSR to
check units required by the different function parameters).
The addWGSR() function will return incorrect values if
the data are not recorded using the required units. For example, this
attempt to add weight-for-age z-scores to the example data:
svy <- addWGSR(data = svy, sex = "sex", firstPart = "weight",
secondPart = "age", index = "wfa")
#> ===========================================================================will give incorrect results:
summary(svy$wfaz)
#> Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
#> 3.450 7.692 9.840 9.684 11.430 15.900 1The odd range of values is due to age being recorded in months rather than days.
It is simple to convert all ages from months to days:
svy$age <- svy$age * (365.25 / 12)
head(svy)
#> psu age sex weight height muac oedema wfhz wfaz
#> 1 1 304.3750 1 5.7 64.2 125 2 -2.73 3.45
#> 2 1 304.3750 2 5.8 64.4 121 2 -2.04 3.95
#> 3 1 273.9375 2 6.5 62.2 139 2 0.13 5.12
#> 4 1 334.8125 9 6.5 64.9 129 2 NA NA
#> 5 1 730.5000 2 6.5 72.9 120 2 -3.44 3.82
#> 6 1 365.2500 2 6.6 69.4 126 2 -2.26 5.01before calculating and adding weight-for-age z-scores:
svy <- addWGSR(data = svy, sex = "sex", firstPart = "weight",
secondPart = "age", index = "wfa")
#> ===========================================================================
head(svy)
#> psu age sex weight height muac oedema wfhz wfaz
#> 1 1 304.3750 1 5.7 64.2 125 2 -2.73 -4.13
#> 2 1 304.3750 2 5.8 64.4 121 2 -2.04 -3.19
#> 3 1 273.9375 2 6.5 62.2 139 2 0.13 -1.97
#> 4 1 334.8125 9 6.5 64.9 129 2 NA NA
#> 5 1 730.5000 2 6.5 72.9 120 2 -3.44 -4.61
#> 6 1 365.2500 2 6.6 69.4 126 2 -2.26 -2.56
summary(svy$wfaz)
#> Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
#> -4.610 -1.873 -1.085 -1.154 -0.480 2.600 1The muac column in the example dataset is recorded in millimetres (mm). We need to convert this to centimetres (cm):
svy$muac <- svy$muac / 10
head(svy)
#> psu age sex weight height muac oedema wfhz wfaz
#> 1 1 304.3750 1 5.7 64.2 12.5 2 -2.73 -4.13
#> 2 1 304.3750 2 5.8 64.4 12.1 2 -2.04 -3.19
#> 3 1 273.9375 2 6.5 62.2 13.9 2 0.13 -1.97
#> 4 1 334.8125 9 6.5 64.9 12.9 2 NA NA
#> 5 1 730.5000 2 6.5 72.9 12.0 2 -3.44 -4.61
#> 6 1 365.2500 2 6.6 69.4 12.6 2 -2.26 -2.56before using the addWGS() function to calculate
MUAC-for-age z-scores:
svy <- addWGSR(svy, sex = "sex", firstPart = "muac",
secondPart = "age", index = "mfa")
#> ===========================================================================
head(svy)
#> psu age sex weight height muac oedema wfhz wfaz mfaz
#> 1 1 304.3750 1 5.7 64.2 12.5 2 -2.73 -4.13 -1.97
#> 2 1 304.3750 2 5.8 64.4 12.1 2 -2.04 -3.19 -1.88
#> 3 1 273.9375 2 6.5 62.2 13.9 2 0.13 -1.97 -0.14
#> 4 1 334.8125 9 6.5 64.9 12.9 2 NA NA NA
#> 5 1 730.5000 2 6.5 72.9 12.0 2 -3.44 -4.61 -2.70
#> 6 1 365.2500 2 6.6 69.4 12.6 2 -2.26 -2.56 -1.46As a last example we will use the addWGSR() function to
add body mass index-for-age (bfa) z-scores to the data to create a new
variable called bmiAgeZ with a precision of 4 decimal places as:
svy <- addWGSR(data = svy, sex = "sex", firstPart = "weight",
secondPart = "height", thirdPart = "age", index = "bfa",
output = "bmiAgeZ", digits = 4)
#> ===========================================================================
head(svy)
#> psu age sex weight height muac oedema wfhz wfaz mfaz bmiAgeZ
#> 1 1 304.3750 1 5.7 64.2 12.5 2 -2.73 -4.13 -1.97 -2.6928
#> 2 1 304.3750 2 5.8 64.4 12.1 2 -2.04 -3.19 -1.88 -2.0005
#> 3 1 273.9375 2 6.5 62.2 13.9 2 0.13 -1.97 -0.14 0.0405
#> 4 1 334.8125 9 6.5 64.9 12.9 2 NA NA NA NA
#> 5 1 730.5000 2 6.5 72.9 12.0 2 -3.44 -4.61 -2.70 -2.8958
#> 6 1 365.2500 2 6.6 69.4 12.6 2 -2.26 -2.56 -1.46 -2.0796To maintain support for earlier versions of the package, the earlier
functions used to calculate anthropometric z-scores for
weight-for-age, height-for-age and
weight-for-height have been kept for now until future
deprecation. For current users, it is recommended to use
addWGSR() and getWGSR() functions.
For this example, we will use the getWGS() function and
apply it to dummy data of a 52 month old male child
with a weight of 14.6 kg and a height of 98.0
cm.
# weight-for-age z-score
waz <- getWGS(sexObserved = 1, # 1 = Male / 2 = Female
firstPart = 14.6, # Weight in kilograms up to 1 decimal place
secondPart = 52, # Age in whole months
index = "wfa") # Anthropometric index (weight-for-age)
waz
#> [1] -1.187651
# height-for-age z-score
haz <- getWGS(sexObserved = 1,
firstPart = 98, # Height in centimetres
secondPart = 52,
index = "hfa") # Anthropometric index (height-for-age)
haz
#> [1] -1.741175
# weight-for-height z-score
whz <- getWGS(sexObserved = 1,
firstPart = 14.6,
secondPart = 98,
index = "wfh") # Anthropometric index (weight-for-height)
whz
#> [1] -0.1790878Applying the getWGS() function results in a calculated
z-score for one child.
For this example, we will use the getCohortWGS()
function and apply it to sample data anthro1 that came with
zscorer.
# Make a call for the anthro1 dataset
anthro1As you will see, this dataset has the 4 variables you will need to
use with getCohortWGS() to calculate the
z-score for the corresponding anthropometric index. These
are age, sex, weight and
height.
head(anthro1)
#> psu age sex weight height muac oedema haz waz whz flag
#> 1 1 6 1 7.3 65.0 146 2 -1.23 -0.76 0.06 0
#> 2 1 42 2 12.5 89.5 156 2 -2.35 -1.39 -0.02 0
#> 3 1 23 1 10.6 78.1 149 2 -2.95 -1.06 0.57 0
#> 4 1 18 1 12.8 81.5 160 2 -0.28 1.42 2.06 0
#> 5 1 52 1 12.1 87.3 152 2 -4.21 -2.68 -0.14 0
#> 6 1 36 2 16.9 93.0 190 2 -0.54 1.49 2.49 0To calculate the three anthropometric indices for all the children in the sample, we execute the following commands in R:
# weight-for-age z-score
waz <- getCohortWGS(data = anthro1,
sexObserved = "sex",
firstPart = "weight",
secondPart = "age",
index = "wfa")
head(waz, 50)
#> [1] -0.75605549 -1.39021503 -1.05597853 1.41575096 -2.67757242
#> [6] 1.49238050 -0.12987704 -0.02348159 -1.50647344 -1.54381630
#> [11] -2.87495712 -0.43497240 -1.03899540 -1.69281855 -1.31245898
#> [16] -2.21003260 -0.01189226 -0.90917762 -0.67839855 -0.94746695
#> [21] -2.49960425 -0.95659644 -1.65442686 -1.25052760 0.67335751
#> [26] 0.30156301 0.24261346 -2.78670709 -1.15820651 -1.15477183
#> [31] -1.35540820 -0.59134959 -4.14967218 -0.45748752 -0.74331669
#> [36] -1.69725836 -1.05745067 -0.18869508 -0.42095770 -2.21030414
#> [41] -1.30536715 -3.63778143 -0.60662526 -0.54360470 -1.59171780
#> [46] -1.74745738 -0.34803338 0.69896149 -0.74467130 0.18924572
# height-for-age z-score
haz <- getCohortWGS(data = anthro1,
sexObserved = "sex",
firstPart = "height",
secondPart = "age",
index = "hfa")
head(haz, 50)
#> [1] -1.2258169 -2.3475886 -2.9518041 -0.2812852 -4.2056663 -0.5387678
#> [7] -2.4020719 -1.0317699 -2.7410884 -4.7037571 -2.5670550 -2.1144960
#> [13] -2.2323505 -2.3155458 -2.7516165 -2.7930694 0.1121349 -1.9001797
#> [19] -2.9543730 -1.9671042 -3.8716522 0.8667206 -2.8252069 -2.1412285
#> [25] -2.7994643 0.5496459 -1.4372002 -3.7979410 -2.5661752 -1.8301183
#> [31] -1.6548589 -2.7110333 -3.6399642 -1.7955069 -1.6775100 -1.0317699
#> [37] -0.4356881 -1.2660152 0.4990326 -4.6085660 -3.1662351 -1.0695930
#> [43] -1.8477936 -2.5502314 -1.8301183 -2.2755493 -3.2816532 0.4876774
#> [49] -2.4396410 -0.4794744
# weight-for-height z-score
whz <- getCohortWGS(data = anthro1,
sexObserved = "sex",
firstPart = "weight",
secondPart = "height",
index = "wfh")
head(whz, 50)
#> [1] 0.05572347 -0.01974903 0.57469112 2.06231749 -0.14080044
#> [6] 2.49047246 1.83315197 0.93614891 0.18541943 2.11599287
#> [11] -1.96943887 1.06351047 0.35315830 -0.61151003 -0.01049441
#> [16] -0.75038993 -0.08000322 0.31277573 1.56456175 0.22152087
#> [21] -0.08798757 -2.14197877 -0.30804823 0.00778227 3.21041413
#> [26] 0.07434468 1.40966986 -0.81485050 0.63816647 -0.33540392
#> [31] -0.61955533 1.35716952 -2.77364671 1.00831095 0.32842063
#> [36] -1.66705281 -1.21157702 0.89024472 -0.89865037 0.82166393
#> [41] 0.64442137 -4.39847850 0.38411140 1.48299847 -0.93068495
#> [46] -0.88558228 1.69551410 0.65143649 0.61269397 0.59813891Applying the getCohortWGS() function results in a vector
of calculated z-scores for all children in the cohort or
sample.
For this example, we will use the getAllWGS() function
and apply it to sample data anthro1 that came with
zscorer.
# weight-for-age z-score
zScores <- getAllWGS(data = anthro1,
sex = "sex",
weight = "weight",
height = "height",
age = "age",
index = "all")
head(zScores, 20)
#> waz haz whz
#> 1 -0.75605549 -1.2258169 0.05572347
#> 2 -1.39021503 -2.3475886 -0.01974903
#> 3 -1.05597853 -2.9518041 0.57469112
#> 4 1.41575096 -0.2812852 2.06231749
#> 5 -2.67757242 -4.2056663 -0.14080044
#> 6 1.49238050 -0.5387678 2.49047246
#> 7 -0.12987704 -2.4020719 1.83315197
#> 8 -0.02348159 -1.0317699 0.93614891
#> 9 -1.50647344 -2.7410884 0.18541943
#> 10 -1.54381630 -4.7037571 2.11599287
#> 11 -2.87495712 -2.5670550 -1.96943887
#> 12 -0.43497240 -2.1144960 1.06351047
#> 13 -1.03899540 -2.2323505 0.35315830
#> 14 -1.69281855 -2.3155458 -0.61151003
#> 15 -1.31245898 -2.7516165 -0.01049441
#> 16 -2.21003260 -2.7930694 -0.75038993
#> 17 -0.01189226 0.1121349 -0.08000322
#> 18 -0.90917762 -1.9001797 0.31277573
#> 19 -0.67839855 -2.9543730 1.56456175
#> 20 -0.94746695 -1.9671042 0.22152087Applying the getAllWGS() function results in a data
frame of calculated z-scores for all children in the cohort
or sample for all the anthropometric indices.
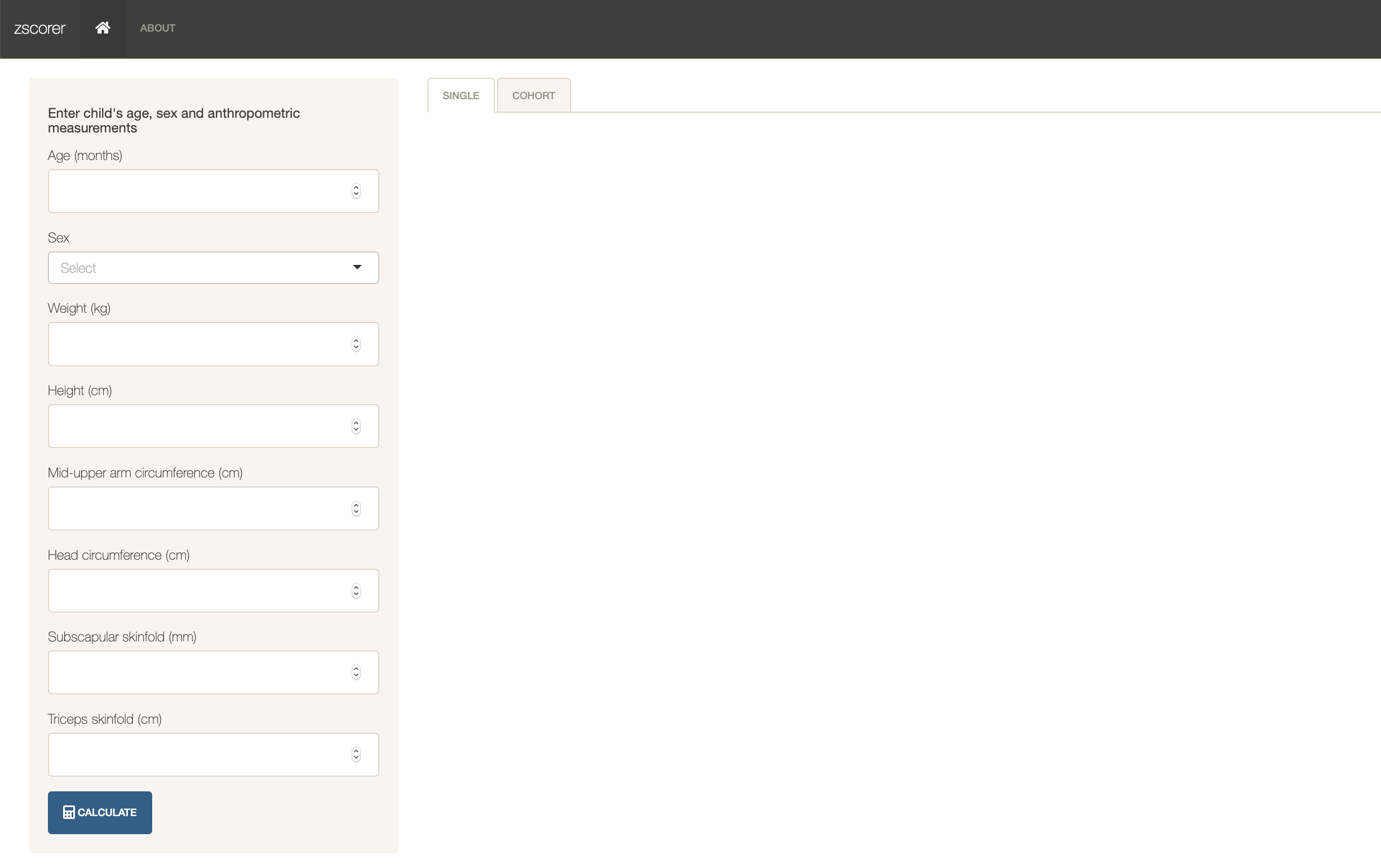
To use the included Shiny app, run the following command in R:
run_zscorer()This will initiate the Shiny app using the installed web browser in your current device as shown below:
World Health Organization. (2006). WHO child growth standards : length/height-for-age, weight-for-age, weight-for-length, weight -for-height and body mass index-for-age : methods and development. World Health Organization. https://apps.who.int/iris/handle/10665/43413
World Health Organization. (2007). WHO child growth standards : head circumference-for-age, arm circumference-for-age, triceps skinfold-for-age and subscapular skinfold-for-age : methods and development. World Health Organization. https://apps.who.int/iris/handle/10665/43706
de Onis M. Development of a WHO growth reference for school-aged children and adolescents. Bull World Health Org. 2007;85: 660–667. doi:10.2471/BLT.07.043497
Mramba L, Ngari M, Mwangome M, Muchai L, Bauni E, Walker AS, et al. A growth reference for mid upper arm circumference for age among school age children and adolescents, and validation for mortality: growth curve construction and longitudinal cohort study. BMJ. 2017;: j3423–8. doi:10.1136/bmj.j3423
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.